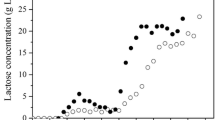
Abstract
In this study, various mathematical functions were utilized to fit the observed values and the kinetic parameters of fermentation in immobilized-cell stirred tank reactor (ICSTR) with carob extract medium (CEM). The best model was selected based on the residual sum of squares (RSS), root-mean-square-error (RMSE), mean-absolute-error (MAE), determination coefficient (R2), slope (m), bias factor (BF), accuracy factor (AF), the objective function (Φ-factor), and f-testing. Therefore, the Stannard (ST) model was good agreement with the actual biomass production (X), ethanol production (P), and sugar consumption (S) data (RSS = 0.22, 6.23, and 45.70 g/L; RMSE = 0.17, 0.88, and 2.39 g/L; MAE = 0.14, 0.64, and 1.72 g/L; R2 = 0.9948, 0.9977, and 0.9972; m = 1.03, 1.02, and 1.02; BF = 1.35, 1.08, and 0.97; AF = 1.41, 1.14, and 1.04; and Φ-factor = 0.008, 0.004, and 0.005 (∑Φ-factor = 0.017) for X, P, and S, respectively). In the prediction of kinetic parameters belonging to the experimental fermentation, the ST model also gave better, satisfactory, and well-directed results than the other mathematical models. Besides, the ST model was also accomplishedly validated using two independent sets of the experimental data yielded from the different carbon sources in various concentrations. Consequently, the models proposed for X, P, and S can serve as universal functions to predict the real values and the kinetic parameters of the ethanol fermentation. Besides, these models can be applied to improve the ethanol fermentation in the ICSTR with CEM.



Similar content being viewed by others
References
Germec M, Kartal F, Bilgic M, Ilgin M, Ilhan E, Güldali H, Isci A, Turhan I (2016) Ethanol production from rice hull using Pichia stipitis and optimization of acid pretreatment and detoxification processes. Biotechnol Prog 32(4):872–882
Germec M, Turhan I (2018) Ethanol production from acid-pretreated and detoxified tea processing waste and its modeling. Fuel 231:101–109
Germec M, Turhan I (2018) Ethanol production from acid-pretreated and detoxified rice straw as sole renewable resource. Biomass Conv Biorefinery 8(3):607–619
Lin S-P, Kuo T-C, Wang H-T, Ting Y, Hsieh C-W, Chen Y-K, Hsu H-Y, Cheng K-C (2020) Enhanced bioethanol production using atmospheric cold plasma-assisted detoxification of sugarcane bagasse hydrolysate. Bioresour Technol 313:123704
Bader NB, Germec M, Turhan I (2020) Scheffersomyces stipitis biofilm reactor for ethanol production from acid-pretreated/detoxified and glucose-or xylose-enriched rice husk hydrolysate under a continuous process. Biomass Conv Bioref. https://doi.org/10.1007/s13399-020-00611-6
Bader NB, Germec M, Turhan I (2021) Ethanol production from different medium compositions of rice husk hydrolysate by using Scheffersomyces stipitis in a repeated-batch biofilm reactor and its modeling. Process Biochem 100:26–38
Germec M, Ozcan A, Turhan I (2019) Bioconversion of wheat bran into high value-added products and modelling of fermentations. Ind Crop Prod 139:111565
Yücel Y, Göycıncık S (2015) Optimization of ethanol production from spent tea waste by Saccharomyces cerevisiae using statistical experimental designs. Biomass Conv Biorefinery 5(3):247–255
Sarris D, Papanikolaou S (2016) Biotechnological production of ethanol: Biochemistry, processes and technologies. Eng Life Sci 16(4):307–329
FAOSTAT (2016) Food and Agriculture Organization of the United Nations. http://faostat3.fao.org/download/O/OA/E (accessed on 16.05.18).
Oziyci HR, Tetik N, Turhan I, Yatmaz E, Ucgun K, Akgul H, Gubbuk H, Karhan M (2014) Mineral composition of pods and seeds of wild and grafted carob (Ceratonia siliqua L.) fruits. Sci Hortic 167:149–152
Lingappa K, Pramod T, Ali SI (2007) Influence of pH on citric acid production by Aspergillus niger under submerged fermentation in carob pod extract. J Sci Ind Res 66(8):618–620
Turhan I, Bialka KL, Demirci A, Karhan M (2010) Enhanced lactic acid production from carob extract by Lactobacillus casei using invertase pretreatment. Food Biotechnol 24(4):364–374
Carvalheiro F, Moniz P, Duarte LC, Esteves MP, Gírio FM (2011) Mannitol production by lactic acid bacteria grown in supplemented carob syrup. J Ind Microbiol Biotechnol 38(1):221–227
Manso T, Nunes C, Raposo S, Lima-Costa ME (2010) Carob pulp as raw material for production of the biocontrol agent P. agglomerans PBC-1. J Ind Microbiol Biotechnol 37(11):1145–1155
Roukas T, Biliaderis CG (1995) Evaluation of carob pod as a substrate for pullulan production by Aureobasidium pullulans. Appl Biochem Biotechnol 55(1):27–44
Oziyci HR, Turhan I, Tetik N, Kulcan AA, Akkoyun T, Yatmaz E, Germec M, Karhan M (2015) Concentration of D-pinitol in carob extract by using multi-stage enrichment processes. J Food 40(3):125–131
Gürler H, Erkan S, Ozcan A, Yilmaze RC, Karahalil E, Germec M, Yatmaz E, Ogel Z, Turhan I (2020) Scale-up processing with different microparticle agent for β-mannanase production in a large-scale stirred tank bioreactor. J Food Process Preserv. https://doi.org/10.1111/jfpp14915
Germec M, Turhan I (2019) Evaluation of carbon sources for the production of inulinase by Aspergillus niger A42 and its characterization. Bioprocess Biosyst Eng 42(12):1993–2005
Ilgın M, Germec M, Turhan I (2020) Statistical and kinetic modeling of Aspergillus niger inulinase fermentation from carob extract and its partial concentration. Ind Crop Prod 156:112866
Turhan I, Bialka KL, Demirci A, Karhan M (2010) Ethanol production from carob extract by using Saccharomyces cerevisiae. Bioresour Technol 101(14):5290–5296
Ercan Y, Irfan T, Mustafa K (2013) Optimization of ethanol production from carob pod extract using immobilized Saccharomyces cerevisiae cells in a stirred tank bioreactor. Bioresour Technol 135:365–371
Germec M, Turhan I, Karhan M, Demirci A (2015) Ethanol production via repeated-batch fermentation from carob pod extract by using Saccharomyces cerevisiae in biofilm reactor. Fuel 161:304–311
Bahry H, Pons A, Abdallah R, Pierre G, Delattre C, Fayad N, Taha S, Vial C (2017) Valorization of carob waste: definition of a second-generation bioethanol production process. Bioresour Technol 235:25–34
Erkan SB, Yatmaz E, Germec M, Turhan I (2020) Effect of furfural concentration on ethanol production using Saccharomyces cerevisiae in an immobilized cells stirred-tank bioreactor with glucose-based medium and mathematical modeling. J Food Process Preserv. https://doi.org/10.1111/jfpp14635
Germec M, Demirci A, Turhan I (2020) Biofilm reactors for value-added products production: an in-depth review. Biocatal Agric Biotechnol 27:101662
Bai F, Anderson W, Moo-Young M (2008) Ethanol fermentation technologies from sugar and starch feedstocks. Biotechnol Adv 26(1):89–105
Germec M, Yatmaz E, Karahalil E, Turhan I (2017) Effect of different fermentation strategies on β-mannanase production in fed-batch bioreactor system. 3. Biotech 7(1):77
Pramod T, Lingappa K (2017) Immobilization of Aspergillus niger in hen egg white for the production of citric acid using carob pod extract. J Microbiol Biotechnol Res 2(2):265–269
Germec M, Turhan I, Demirci A, Karhan M (2016) Effect of media sterilization and enrichment on ethanol production from carob extract in a biofilm reactor. Energy Sources, Part A: Recovery, Utilization, and Environmental Effects 38(21):3268–3272
Tokuyama K, Shimodaira Y, Terawaki T, Kusunose Y, Nakai H, Tsuji Y, Toya Y, Matsuda F, Shimizu H (2020) Data science-based modeling of the lysine fermentation process. J Biosci Bioeng 130(4):409–415
Baranyi J, Roberts T (1994) A dynamic approach to predicting bacterial growth in food. Int J Food Microbiol 23(3-4):277–294
Morgan P, Mercer L, Flodin N (1975) General model for nutritional responses of higher organisms. Proc Natl Acad Sci 72(11):4327–4331
Stannard C, Williams A, Gibbs P (1985) Temperature/growth relationships for psychrotrophic food-spoilage bacteria. Food Microbiol 2(2):115–122
Weibull W (1951) A statistical distributipon function of wide applicability. J Appl Mech 9:293–297
Zwietering M, Jongenburger I, Rombouts F, Van't Riet K (1990) Modeling of the bacterial growth curve. Appl Environ Microbiol 56(6):1875–1881
Lee KH, Choi IS, Kim Y-G, Yang D-J, Bae H-J (2011) Enhanced production of bioethanol and ultrastructural characteristics of reused Saccharomyces cerevisiae immobilized calcium alginate beads. Bioresour Technol 102(17):8191–8198
Instruments Y-YS (2009) YSI 2700 SELECT Biochemistry analyzer-user’s manual. Yellow Springs, Ohio
Miller G (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31(3):426–428
Germec M, Cheng K-C, Karhan M, Demirci A, Turhan I (2020) Application of mathematical models to ethanol fermentation in biofilm reactor with carob extract. Biomass Conv Biorefinery 10:237–252
Germec M, Turhan I, Karhan M, Demirci A (2019) Kinetic modeling and techno-economic feasibility of ethanol production from carob extract based medium in biofilm reactor. Appl Sci 9(10):2121
Chai T, Draxler R (2014) Root mean square error (RMSE) or mean absolute error (MAE)?–arguments against avoiding RMSE in the literature. Geosci Model Dev 7(3):1247–1250
Cheng K, Demirci A, Catchmark J, Puri V (2010) Modeling of pullulan fermentation by using a color variant strain of Aureobasidium pullulans. J Food Eng 98(3):353–359
Cheng K-C, Demirci A, Catchmark JM, Puri VM (2011) Effects of initial ammonium ion concentration on pullulan production by Aureobasidium pullulans and its modeling. J Food Eng 103(2):115–122
Ross T (1996) Indices for performance evaluation of predictive models in food microbiology. J Appl Microbiol 81(5):501–508
Feng J, Zhang J-S, Jia W, Yang Y, Liu F, Lin C-C (2014) An unstructured kinetic model for the improvement of triterpenes production by Ganoderma lucidum G0119 based on nitrogen source effect. Biotechnol Bioprocess Eng 19(4):727–732
Lomax RG, Hahs-Vaughn DL (2013) An introduction to statistical concepts. Routledge,
Germec M, Karhan M, Demirci A, Turhan I (2020) Implementation of flexible models to bioethanol production from carob extract–based media in a biofilm reactor. Biomass Conv Biorefinery. https://doi.org/10.1007/s13399-020-00612-5
Erkan SB (2020) Investigation of the effect of inhibitors on ethanol production with immobilized Saccharomyces cerevisiae cells in calcium alginate. Master Thesis, Akdeniz University, Antalya, Turkey
Yatmaz E (2012) Optimization of ethanol production from carob pod extract by using Saccharomyces cerevisiae cells immobilized in Ca-alginate. Master Thesis, Akdeniz University, Antalya, Turkey
Lübken M, Gehring T, Wichern M (2010) Microbiological fermentation of lignocellulosic biomass: current state and prospects of mathematical modeling. Appl Microbiol Biotechnol 85(6):1643–1652
Ilgın M, Germec M, Turhan I (2020) Inulinase production and mathematical modeling from carob extract by using Aspergillus niger. Biotechnol Prog 36(1):e2919
Germec M, Karhan M, Demirci A, Turhan I (2020) Mathematical modeling of batch bioethanol generation from carob extract in the suspended-cell stirred-tank bioreactor. Int J Energy Res 44(11):9021–9034
Sapra R (2014) Using R2 with caution. Curr Med Res Pract 4(3):130–134
Kissell R, Poserina J (2017) Regression models. In: Kissell R, Poserina J (eds) Optimal Sports Math, Statistics, and Fantasy. Academic Press, pp 39-67. doi:https://doi.org/10.1016/B978-0-12-805163-4.00002-5
Baranyi J, Pin C (2004) Modeling the history effect on microbial growth and survival: deterministic and stochastic approaches. In: Modeling Microbial Responses in Food. CRC Press Inc. Boca Raton, pp 285-302
Germec M, Karhan M, Bialka K, Demirci A, Turhan I (2018) Mathematical modeling of lactic acid fermentation in bioreactor with carob extract. Biocatal Agric Biotechnol 14:254–263
Gürler HN, Germec M, Turhan I (2020) The inhibition effect of phenol on the production of Aspergillus niger inulinase and its modeling. J Food Process Preserv. https://doi.org/10.1111/jfpp14522
Karahalil E, Germeç M, Turhan I (2019) β-mannanase production and kinetic modeling from carob extract by using recombinant Aspergillus sojae. Biotechnol Prog 35(6):e2885
Funding
This study was funded by the Akdeniz University Research Foundation (grant number 2012.02.0121.012).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 16.9 kb)
Rights and permissions
About this article
Cite this article
Yatmaz, E., Germec, M., Erkan, S.B. et al. Modeling of ethanol fermentation from carob extract–based medium by using Saccharomyces cerevisiae in the immobilized-cell stirred tank bioreactor. Biomass Conv. Bioref. 12, 5241–5255 (2022). https://doi.org/10.1007/s13399-020-01154-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13399-020-01154-6