Abstract
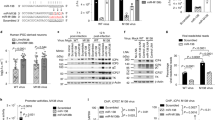
During latent infection, the HSV-1 virus generates only a single transcript, LAT, which encodes six miRNAs. The GABAergic pathway signaling system is an essential cell signaling pathway influenced by various therapeutic targets and some brain disorders, such as epilepsy. This study found that miRNAs encoding LAT might target the STXBP1 and GABBR2 genes, which are among the significant genes in the GABAergic pathway. Bioinformatic analysis utilizing TargetScan version 5.2 and the RNA22 tools uncovered miRNAs encoding LAT that can influence STXBP1 and GABBR2 transcripts. To evaluate the targeting effect of candidate microRNAs encoding LAT, namely, miR-H3 and miR-H4, LAT constructs were transfected into HEK 293T cells. The expression levels of microRNAs encoding LAT, as well as STXBP1 and GABBR2, were assayed by real-time PCR. Finally, the targeting potential of STXBP1 and GABBR2 3′UTR by LAT-encoded microRNAs was evaluated by the luciferase assay. In the current study, the bioinformatic tool TargetScan demonstrated that miR-H3 has the potential to target the transcripts of the STXBP1 and GABBR2 genes, whereas miR-H4 solely targeted GABBR2. On the other hand, the bioinformatic tool RNA22 validated the potential targeting of STXBP1 and GABBR2 by miR-H3 and miR-H4. Our findings showed that overexpression of miR-H4, miR-H3, or LAT significantly decreased STXBP1 gene expression by an average of 0.0593-fold, 0.237-fold, and 0.84-fold, respectively. Similarly, overexpression of miR-H3 or miR-H4 decreased GABBR2 expression by an average of 0.055- or 0.687-fold, respectively. Notably, targeting the GABBR2 3′UTR with the LAT transcript had no detectable effect. The evaluation of the targeting potential of STXBP1 and GABBR2 3′UTR by microRNAs encoded by LAT was conducted with a luciferase assay. Our results showed that miR-H3 overexpression reduces Renilla expression in psiCHECK2 plasmids with STXBP1 or GABBR2 3′UTR genes by 0.62- and 0.55-fold, respectively. miR-H4 reduced Renilla gene expression regulated by GABBR2’s 3′UTR plasmid but had no effect on the Renilla gene expression regulated by STXBP1’s 3′UTR. When the LAT transcript was overexpressed, there was a decrease in Renilla expression by 0.44-fold because of the regulation of STXBP1’s 3′UTR. However, there was no significant effect observed through the control of GABBR2’s 3′UTR.


Similar content being viewed by others
Data availability
The data utilized in this study have been archived and are accessible to the public upon request by the corresponding author. Additionally, the data can be found at the Department of Microbiology, which is located within the School of Biology at the College of Science, University of Tehran.
Code availability
The software used in this study is commercially available. No custom programming has been carried out for this project.
References
Arefian E, Soleimani M, Bamdad T, Mobarra Z, Hajarizadeh A (2011) Herpes simplex virus type 1 latency-associated transcript reduces human neuroblastoma cell proliferation
Boss IW, Plaisance KB, Renne R (2009) Role of virus-encoded microRNAs in herpesvirus biology. Trends Microbiol 17(12):544–553
Choi S, Lee S, Han Y, Choi J, Kim I, Lee J, An H (2022) miR-31–3p functions as a tumor suppressor by directly targeting GABBR2 in prostate cancer. Front Oncol 12
Christensen M, Schratt GM (2009) microRNA involvement in developmental and functional aspects of the nervous system and in neurological diseases. Neurosci Lett 466(2):55–62
Cid MP, Vilcaes AA, Rupil L, Salvatierra NA, Roth GA (2011) Participation of the GABAergic system on the glutamate release of frontal cortex synaptosomes from Wistar rats with experimental autoimmune encephalomyelitis. Neuroscience 189:337–344
Duarte LF, Farías MA, Álvarez DM, Bueno SM, Riedel CA, González PA (2019) Herpes simplex virus type 1 infection of the central nervous system: insights into proposed interrelationships with neurodegenerative disorders. Front Cell Neurosci 13:46
Farrant M, Nusser Z (2005) Variations on an inhibitory theme: phasic and tonic activation of GABAA receptors. Nat Rev Neurosci 6(3):215–229
Farrell MJ, Dobson AT, Feldman LT (1991) Herpes simplex virus latency-associated transcript is a stable intron. Proc Natl Acad Sci 88(3):790–794
Froestl W (2011) An historical perspective on GABAergic drugs. Future Med Chem 3(2):163–175
Gengyo-Ando K, Kitayama H, Mukaida M, Ikawa Y (1996) A murine neural-specific homolog corrects cholinergic defects in Caenorhabditis elegans unc-18 mutants. J Neurosci 16(21):6695–6702
Held K, Derfuss T (2011) Control of HSV-1 latency in human trigeminal ganglia—current overview. J Neurovirol 17(6):518–527
Hogestyn JM, Mock DJ, Mayer-Proschel M (2018) Contributions of neurotropic human herpesviruses herpes simplex virus 1 and human herpesvirus 6 to neurodegenerative disease pathology. Neural Regen Res 13(2):211
Hu D, Wang Y, Zhang H, Kong D (2018) Identification of miR-9 as a negative factor of insulin secretion from beta cells. J Physiol Biochem 74(2):291–299
Ikebuchi Y, Kanda T, Ikeda H, Yoshida A, Sakaguchi T, Urabe S, Minami H, Nakao K, Kuwamoto S, Inoue H (2020) Identification of human herpes virus 1 encoded micro RNA s in biopsy samples of lower esophageal sphincter muscle during peroral endoscopic myotomy for esophageal achalasia. Dig Endosc 32(1):136–142
Jeshvaghani ZS, Arefian E, Asgharpour S, Soleimani M (2021) Latency-associated transcript-derived microRNAs in herpes simplex virus type 1 target SMAD3 and SMAD4 in TGF-β/Smad signaling pathway. Iran Biomed J 25(3):169
Jin L, Perng G-C, Mott KR, Osorio N, Naito J, Brick DJ, Carpenter D, Jones C, Wechsler SL (2005) A herpes simplex virus type 1 mutant expressing a baculovirus inhibitor of apoptosis gene in place of latency-associated transcript has a wild-type reactivation phenotype in the mouse. J Virol 79(19):12286–12295
Katsu M, Hama Y, Utsumi J, Takashina K, Yasumatsu H, Mori F, Wakabayashi K, Shoji M, Sasaki H (2019) MicroRNA expression profiles of neuron-derived extracellular vesicles in plasma from patients with amyotrophic lateral sclerosis. Neurosci Lett 708:134176
Khaikin Y, Mercimek-Andrews S (2016) STXBP1 encephalopathy with epilepsy. GeneReviews®[Internet]
Kodera H, Kato M, Nord AS, Walsh T, Lee M, Yamanaka G, Tohyama J, Nakamura K, Nakagawa E, Ikeda T (2013) Targeted capture and sequencing for detection of mutations causing early onset epileptic encephalopathy. Epilepsia 54(7):1262–1269
Lang H, Ai Z, You Z, Wan Y, Guo W, Xiao J (2015) Characterization of miR-218/322. Endocrinology 54:65–73
Olson HE, Tambunan D, LaCoursiere C, Goldenberg M, Pinsky R, Martin E, Ho E, Khwaja O, Kaufmann WE, Poduri A (2015) Mutations in epilepsy and intellectual disability genes in patients with features of Rett syndrome. Am J Med Genet A 167(9):2017–2025
Otsuka M, Oguni H, Liang JS, Ikeda H, Imai K, Hirasawa K, Imai K, Tachikawa E, Shimojima K, Osawa M (2010) STXBP1 mutations cause not only Ohtahara syndrome but also West syndrome—result of Japanese cohort study. Epilepsia 51(12):2449–2452
Rezazadeh A, Uddin M, Snead OC III, Lira V, Silberberg A, Weiss S, Donner EJ, Zak M, Bradbury L, Scherer SW (2019) STXBP1 encephalopathy is associated with awake bruxism. Epilepsy Behav 92:121–124
Romaniello R, Saettini F, Panzeri E, Arrigoni F, Bassi MT, Borgatti R (2015) A de-novo STXBP1 gene mutation in a patient showing the Rett syndrome phenotype. NeuroReport 26(5):254–257
Saitsu H, Kato M, Mizuguchi T, Hamada K, Osaka H, Tohyama J, Uruno K, Kumada S, Nishiyama K, Nishimura A (2008) De novo mutations in the gene encoding STXBP1 (MUNC18-1) cause early infantile epileptic encephalopathy. Nat Genet 40(6):782–788
Scaria V, Hariharan M, Maiti S, Pillai B, Brahmachari SK (2006) Host-virus interaction: a new role for microRNAs. Retrovirology 3(1):1–9
Tang S, Bertke AS, Patel A, Wang K, Cohen JI, Krause PR (2008) An acutely and latently expressed herpes simplex virus 2 viral microRNA inhibits expression of ICP34. 5, a viral neurovirulence factor. Proc Nat Acad Sci 105(31):10931–10936
Thompson R, Sawtell N (2000) HSV latency-associated transcript and neuronal apoptosis. Science 289(5485):1651–1651
Tiwari D, Peariso K, Gross C (2018) MicroRNA-induced silencing in epilepsy: opportunities and challenges for clinical application. Dev Dyn 247(1):94–110
Uğurel E, Şehitoğlu E, Tüzün E, Kürtüncü M, Çoban A, Vural B (2016) Increased complexin-1 and decreased miR-185 expression levels in Behçet’s disease with and without neurological involvement. Neurol Sci 37(3):411–416
Umbach JL, Kramer MF, Jurak I, Karnowski HW, Coen DM, Cullen BR (2008) MicroRNAs expressed by herpes simplex virus 1 during latent infection regulate viral mRNAs. Nature 454(7205):780–783
Umbach JL, Nagel MA, Cohrs RJ, Gilden DH, Cullen BR (2009) Analysis of human alphaherpesvirus microRNA expression in latently infected human trigeminal ganglia. J Virol 83(20):10677–10683
Umbach JL, Wang K, Tang S, Krause PR, Mont EK, Cohen JI, Cullen BR (2010) Identification of viral microRNAs expressed in human sacral ganglia latently infected with herpes simplex virus 2. J Virol 84(2):1189–1192
Wang J, Zhao J (2021) MicroRNA dysregulation in epilepsy: from pathogenetic involvement to diagnostic biomarker and therapeutic agent development. Front Mol Neurosci 14:650372
Yoo Y, Jung J, Lee YN, Lee Y, Cho H, Na E, Hong J, Kim E, Lee JS, Lee JS (2017) GABBR2 mutations determine phenotype in Rett syndrome and epileptic encephalopathy. Ann Neurol 82(3):466–478
Zhu M, Wang L, Zhu J, Xu H, Wei K, Chen Q, Wu X, Miao X, Lu Z (2020) MicroRNA-330 directs downregulation of the GABABR2 in the pathogenesis of pancreatic cancer pain. J Mol Neurosci 70(10):1541–1551
Acknowledgements
The authors would like to acknowledge Dr. Zahra Fekrirad for providing technical assistance.
Funding
The Iran National Science Foundation (INSF) funded this research (grant number 99019844). The decision to publish this article, as well as the study’s design and data collection, analysis, and interpretation, was all made independently of the funder.
Author information
Authors and Affiliations
Contributions
K. M. AL-Khfaji and Nika K. Zamani performed the experimental procedures, data analysis, and manuscript drafting. Ehsan Arefian conceptualized the study and revised the manuscript. The present study is under the supervision of Dr. Ehsan Arefian, who serves as the guarantor of the work. Dr. Arefian has unrestricted access to all the data collected during the study and is accountable for ensuring the integrity and precision of the data analysis. The authors assert that all data were produced internally and that no external sources were utilized.
Corresponding author
Ethics declarations
Ethics approval
The study was carried out in accordance with approved protocols from the Department of Microbiology, School of Biology, College of Science, University of Tehran.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Key points
• STXBP1 is a protein expressed in neuronal cells and has a crucial role in docking and fusing synaptic vesicles.
• Gene expression assays and luciferase assays showed that HSV-1 LAT reduced STXBP1 gene expression by 0.84- and 0.44-fold, respectively.
• miR-H3 directly targets the 3′UTR of STXBP1 and GABBR2, resulting in a decrease in transcript levels of these genes.
• Real-time PCR analysis showed that miR-H4 decreased STXBP1 and GABBR2 gene expression. Luciferase assays showed a significant difference only when miR-H4 targeted the 3′UTR of the GABBR2 gene but not when it targeted the STXBP1 gene.
•The miR-H3 and miR-H4 microRNAs from HSV-1 may contribute to the dysfunction in the GABAergic pathway signaling system, which is linked to neurological diseases.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
AL-Khfaji, K.M.S., Zamani, N.K. & Arefian, E. HSV-1 latency-associated transcript miR-H3 and miR-H4 target STXBP1 and GABBR2 genes. J. Neurovirol. 29, 669–677 (2023). https://doi.org/10.1007/s13365-023-01174-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13365-023-01174-8