Abstract
The aggregation of Cu,Zn-superoxide dismutase (SOD1) plays an important role in the etiology of amyotrophic lateral sclerosis (ALS). For the disruption of ALS progression, discovering new drugs or compounds that can prevent SOD1 aggregation is important. In this study, ESI-MS was used to investigate the interaction of catechins and SOD1. The noncovalent complex of catechins that interact with SOD1 was found and retained in the gas phase under native ESI-MS condition. The conformation changes of SOD1 after binding with catechins were also explored via traveling wave ion mobility (IM) spectrometry. Epigallocatechin gallate (EGCG) can stabilize SOD1 conformation against unfolding in three catechins. To further evaluate the efficacy of EGCG, we monitored the fluorescence changes of dimer E2,E2,-SOD1(apo-SOD1, E:empty) with and without ligands under denaturation conditions, and found that EGCG can inhibit apo-SOD1 aggregation. In addition, the circular dichroism spectra of the samples showed that EGCG can decrease the β-sheet content of SOD1, which can produce aggregates. These results indicated that orthogonal separation dimension in the gas-phase IM coupled with ESI-MS (ESI-IM-MS) can potentially provide insight into the interaction between SOD1 and small molecules. The advantage is that it dramatically decreases the analysis time. Meantime, optical spectroscopy techniques can be used to confirm ESI-IM-MS results.

ᅟ
Similar content being viewed by others
Introduction
Amyotrophic lateral sclerosis (ALS) is a paralytic disorder clinically characterized by the degeneration of motor neurons in the brain and spinal cords. Previous studies have shown that the structure defects of Cu,Zn-superoxide dismutase (SOD1) are associated with ALS [1]. The pathology mechanisms of sporadic and familial ALS caused by SOD1 misfolding and aggregation have been the focus of intense study [2, 3]. Relevant evidence suggests that ALS may be interrelated with SOD1 misfolding induced by metal depletion, disruption of the quaternary structure, or oxidation [4]. Thus, discovering bioactive compounds that can suppress the abnormal oligomerization of SOD1 to ameliorate and treat this disease is urgently necessary [5]. Some researches have confirmed that catechins possess many physiological functions, such as antihypertensive effect, antifibrotic property, and neuroprotective capacity [6]. Epigallocatechin gallates (EGCG) in catechins is mainly polyphenol. A study of the potential protective effects of EGCG in the G93A SOD mouse model of ALS had been reported [7]. Meantime, EGCG can efficiently inhibit amyloid fibrillogenesis in some neurodegenerative disorders, thereby preventing the misfolding proteins or polypeptides to form toxic and on-pathway aggregation intermediates. Therefore, EGCG is a promising agent to modulate fibril maturity [8,9,10,11].
Advanced analytical techniques, such as MS, NMR, and optical spectroscopy are essential to the investigation of the interactions between proteins and small molecules [12,13,14]. ESI-MS has a wider range of applicability in structural biology because of its rapidity, sensitivity, and specifity for biomolecule detection [15, 16]. As a useful tool for study of noncovalent interactions of protein and small molecules, ESI-MS can provide the binding stoichiometry and specificity of noncovalent interaction with rapid and sensitive features [17]. ESI-MS method can also provide insight into the gas-phase stability through complex Vc50 (defined as the cone voltage resulting in 50% dissociation of a complex) determination, further validating the relative binding affinities of different ligands [18]. Meantime, ion mobility (IM) combined with ESI-MS (ESI-IM-MS) is well suited to the study of protein misfolding. The general principle of IM depends on the mobility behaviors of gas-phase ions, and such behaviors are based on the collision cross-sections of these ions. Thus, mobility behavior can be used to identify and characterize protein–protein or protein–ligand interactions [19, 20]. It can also yield significant structural information on proteins and reveal the mechanistic details pertaining to the oligomerization of amyloid disorders on a millisecond time scale [21,22,23]. Meanwhile, collision-induced unfolding (CIU) is a novel methodology. Its principle is as follows: the unfolded forms of protein are generated by collision heating in the gas phase and subsequently separated through IM. The unfolded forms are then detected by TOF mass spectrometry. This method is often used to explore the subtle differences and conformational transitions between the protein and noncovalent complex of proteins and small molecules [24, 25]. ESI-IM-MS has been used in several studies on structural biology. Han et al. [26, 27] reported that bound cation/anion can stabilize multiprotein complexes through IM-MS. Zhao et al. [28] investigated the gas-phase conformation changes of protein–ligand complex by traveling wave ion mobility-mass spectrometry (TWIMS). Tian et al. [29] introduced a new approach for the characterization of disulfide bonding pattern, glycosylation, and antibody structure through IM-MS.
In this report, we employed ESI-MS and collision-induced dissociation (CID) experiment to evaluate the binding affinities of ligands and determine the Vc50 values of protein-ligand system. In this experiment, the metalation states of SOD1 were probed by ESI-MS, in agreement with previous report [30], and three catechins, ([EGCG], epigallocatechin [EGC], and epicatechingallate [ECG]) with similar polyphenol structures as ligands were discussed and analyzed. The effects of ligand binding on the conformational changes of SOD1 were observed by collision induced unfolding (CIU) under different collision energies. Fluorescence experiments were performed to test the effect of EGCG on the aggregation of apo-SOD1 proteins under denatured condition. Furthermore, as a complemental experiment, far UV circular dichroism was used to observe the influence of EGCG on β-sheet content.
Experimental Details
Materials
Bovine SOD1 was purchased from Beyotime Biotechnology (Shanghai, China). The three catechins (EGCG, ECG, and EGC) and ammonium acetate (NH4OAC) were purchased from Sigma-Aldrich (St. Louis, MO, USA). Methanol (chromatographic grade CH3OH) was purchased from TEDIA Company (Fairfield, OH, USA). Trifluoroethanol (TFE), thioflavin-T(ThT), and EDTA were purchased from J&K Scientific Ltd. (Beijing, China). Ammonium 8-anilino-1-naphthalenesulfonate (ANS-NH4) was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). The MicroFloat-A-Lyzer (MW-cut: 10 kDa) used in the dialysis experiment was purchased from Spectrum Laboratories Inc. (Shanghai, China). Ultrapure water was prepared by Milli-Q (Millipore, Bedford, MA, USA).
Sample Preparation
The protein sample was dissolved into 10 mM of NH4OAC to a stock concentration of 100 μM. The ligand samples were dissolved to 3 mM with 10% CH3OH, and the resulting solution was further diluted to 300 μM with ultrapure water. Before the experiment, the protein and ligand stock solutions were stored at –20 and 4 °C. Binding experiments of SOD1 with catechins (EGCG/EGC/ECG) were performed in a protein:ligand ratio of 1:6 after the mixtures were incubated at 37 °C for 1 h.
Preparations of apo-SOD1
Apo-dimer SOD1 was prepared using a demetallation protocol [31]. Briefly, 1 mL of stock solution was dialyzed against 20 mM NH4OAC buffer (pH 3.2), which contains 5 mM EDTA, for 24 h in a dialysis system. The dialysis solution was then changed to 20 mM NH4OAC buffer (pH 3.2) without 5 mM EDTA. After 24 h, the buffer was replaced with 20 mM NH4OAC (pH 6.8), and resulting solution was dialyzed for 24 h. The metal content of the apo-SOD1 sample was determined through ESI-MS analysis. Supplementary Figure S1 shows the ESI-MS spectrum and deconvolution mass spectrum of apo-SOD1, the average molecular mass of apo-SOD1 is 31183.2Da, which is consistent with previous reports [30]. The apo- SOD1 sample was stored at –20 °C.
Binding Affinity Assessment by ESI-IM-MS and MS/MS Experiments
Binding experiments were carried out on a quadrupole-IM-TOF mass spectrometer with ESI source (Synapt G2 HDMS, Waters, Milford, MA, USA). Data were acquired in positive ion mode. The MS tune parameters were carefully adjusted to improve sensitivity. Optimized parameters were set as follows: 20 V cone voltage, 2.35 kV capillary voltage, 80 °C source temperature, 3 V extraction cone voltage, 30 L h.-1 cone gas flow, 450 L h-1 desolvation gas flow rate, 150 °C temperature, 5 μL min-1 sample flow rate. Data were acquired and processed by Masslynx4.0 (Waters, Manchester, UK). The ESI-MS spectra were deconvoluted using MaxEnt software (Waters Corp., Manchester, UK). For IM-MS experiments, the traveling wave height and wave velocity were set to 40 V and 900 ms-1, respectively, and the IMS data were processed by the driftscope software (Waters Corp., Manchester, UK).
Vc50 Experiment
SOD1 and ligand (1:6) were incubated at 37 °C for 1 h. Dissociation induced by in-source collision was achieved by increasing the cone voltage from 10 to 90 V at 4 V increments. Each experiment was performed in triplicate. The 1:1 complex population, deduced from relative intensities of the +11 charge state, was plotted as a function of cone voltage, and Vc50 is the cone voltage value at which 50% of the complex is dissociated.
Collision Induced Unfolding (CIU) Experiment
The gas-phase transition of the selected ion state was monitored through a Synapt G2 mass spectrometer coupled with MS/MS model. A trap collision energy of 20–38 V was applied to the selected ions in the trap cell before the IM separation, in which the selected ions corresponded to the +11 charge state of protein and different complex dimers. A cone voltage of 20 V was set to ensure the signal intensity. IM wave velocity and height were adjusted to 1100 m s-1 and 40 V, respectively. IM data were recorded and processed by MassLynx 4.0. CIU fingerprint was deduced by Origin Pro 2015. The IMS spectra of the selected ions were compared at each collision voltage to determine the changes in the conformations of the proteins and different complex dimers.
Thioflavin T (ThT) Fluorescence Assay
TFE is a protein denaturing agent used extensively to induce proteins to aggregate into fibril-like amyloid. Apo-SOD1 aggregation was investigated by using TFE at 5.5 pH. The amyloid states of the TFE-induced apo-SOD1 were monitored through the changes in the fluorescence spectra of ThT, which can bind to the amyloid fibril. The kinetics data of TFE-induced fibril was recorded by a molecular device (SpectraMaxi3x). Each well in a black Costar 96-well microplate contained 10 μL of protein, 24 μL of TFE, and 10 μL of ThT stock solution in NH4OAC buffer. EGCG of different moral ratios were added to the 96-well microplate. The control had no TFE. The plates were incubated at 37 °C before reading, and the fluorescence spectra were measured every 2 min at excitation and the emission wavelength of 450 and 485 nm, respectively.
Anilinonaphthalene-8-Sulfonic Acid (ANS) Fluorescence Assay
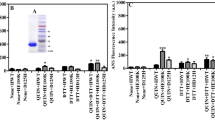
The hydrophobic exposure of the protein was estimated through ANS modification. ANS binds to the hydrophobic parts of protein by hydrophobic interaction. Before the experiment, the sample solutions were incubated at 25 °C for 30 min without denaturant TFE. Spectra Max i3x was used to measure 200 μL of the reaction solution, which contained 0.1 mg of apo-SOD1, 0.013 mg/0 mg of EGCG, 12% TFE, and 4.7 ng of ANS. The control experiment was performed using ANS and EGCG natural solution. When the sample was incubated with TFE for 2 h, the fluorescence emission spectra (Figure 7) were observed at 370 nm excitation wavelength in Spectra Maxi 3x.
Circular Dichroism (CD) Assay
Apo-SOD1 (100 μM) was incubated with EGCG with molar ratio of 1:10 nearly at physiological conditions and then without EGCG. The samples were diluted to 10 μM in 10 mM NH4OAC. Control samples were incubated in parallel. One of the controls contained EGCG but no apo-SOD1, whereas the other control only contained buffer solution. The CD spectra of apo-SOD1 were measured from 200 to 250 nm on a JASCO J-810 spectropolarimeter (Tokyo, Japan) with a computer-controlled water bath. The optical chamber of CD spectrometer was deoxygenated with dry purified nitrogen (99.99%) for 45 min before use and under maintained nitrogen atmosphere. Three scans were obtained and automatically averaged after subtracting the background spectrum. The CD spectra were processed and smoothed using Origin pro 2015.
Results and Discussion
Comparison of the Binding Affinities of Catechins by ESI-IM-MS, MS/MS and Vc50 Experiments
Relative Binding Affinities Assessment of Three Ligands by ESI-IM-MS and MS/MS Experiments
The dominate dimer(Di) and monomer(Mo) species of SOD1 were observed through ESI-MS under “near-native” condition, and the observed charge states ranged from +10 to +13; the charge distribution of a small amount monomer is from +5 to +8. The average molecular mass of SOD1 is 31432.5 Da obtained from Supplementary Figure S1 by using MaxEnt software. In agreement with the previous studies [31, 32], the overlapping peak of ligand mono-adducts on monomer and di-adducts on dimer make the relative affinity assessment complicated. We can evaluate the relative affinity by IM-MS from the IM-MS Driftscope plots. Take EGCG-SOD1 complex for example; the dimeric and monomeric of SOD1 species can be distinguished easily (Figure 2b), so we can extract and export the mass spectrum (Figure 2c) only containing the dimeric species of SOD1 by the driftscope software, and evaluate the relative affinity of different ligands. Figure 1 shows the structures of the three ligands, and Supplementary Figure S2B shows the different binding stoichiometries after incubation with the three ligands. Mainly 1:1 and 1:2 binding stoichiometries of SOD1:ligand were observed at 1:6 protein:ligand ratio after co-incubation at 37 °C for 1 h. However, a nonspecific binding was observed after excessive ligand addition. The equation \( \frac{I\left(1:n\right)}{I(P)+I\left(1:n\right)} \) was applied to determine the relative binding affinities of the three ligands, where P and 1:n were defined as free dimer SOD1 and the different binding stoichiometries of SOD1 and ligand, I (P) and I(1:n) represent the relative abundance of the each charge states of free dimer and SOD1–ligand complexes. The relative intensities of the proteins or complexes in the mass spectra are assumed to be proportional to their relative abundances in the solution [33, 34]. The relative ligand affinity of EGCG, ECG, and EGC binding to SOD1 can be calculated from their extractive mass spectra.The relative affinities of EGCG/ECG/EGC ligand to SOD1 deduced by the equation is 0.38/0.35/0.36, respectively. The ligand’s hydroxyl groups play an important role in interacting with amino acids in proteins via hydrogen bonding. However, we found the relative binding affinity of EGC, which has the least hydroxyl groups, to be a little higher than the ECG, so speculated that EGC has a stronger nonspecific binding. MS/MS experiment was done in order to test our guess. The precursor ions of EGCG /ECG /EGC-SOD1 complex at m/z 2900/2899/2886 were selected and subjected to various trap energies. A comparison of the ESI-MS/MS spectra suggested that SOD1-EGC start to dissociate in a lower energy. In contrast, the SOD1-EGCG and SOD1-ECG have little dissociation at the same energy. Besides, the SOD1-EGC complex almost completely dissociates at the trap energy of 35 V; however, when the trap energy is at 40 V, the dissociation of 48% SOD1-EGCG complex and 55% SOD1-ECG complex was observed (Supplementary Figure S3). The result of MS/MS experiment shows the order of binding forces is EGCG>ECG>EGC.
Native mass spectra of noncovalent complexes of SOD1 and EGCG at molar ratio 1:6 of SOD1:ligand (a), IMS-MS driftscope plots of SOD1 and SOD1-EGCG complex total species (b), the extracted mass spectra of the dimeric SOD1 and SOD1-EGCG complex species from the 2D IM-MS plots (c). Monomeric and dimeric SOD1 species are circled with white and green ellipse and labeled as Mo and Di, respectively. Peaks marked with one circle represent the 1:1 complex, those with two circles represent the 1:2 complex, and those with three circles represent the 1:3 complex
Drift time distributions of the 11+ charge state of SOD1 dimer and its complexes. SOD1 dimer (P, black traces) and SOD1 dimer–EGCG complex (1:1 (PL,red traces), and 1:2 (PL2,green traces) at different trap collision voltages(20, 34, 36 and 38 V, three different conformations, the compact/extended/more extended conformation were labeled as A/B/C, respectively
Influence of EGCG on TFE-induced apo-SOD1 fibril formation. Fibril formation kinetics of apo-SOD1 with different EGCG molar ratios were monitored by ThT fluorescence. Apo-SOD1:EGCG at 1:0 (black), 1:1(red), 1:6 (blue),1:10 (purple), and control (brown). Control sample is the solution of apo-SOD1 protein without TFE
Confirming the Order of Ligand Binding Affinities through Vc50 Measurement
The gas-phase stability of the protein–ligand complex can be assessed by using the Vc50 values. The progressive dissociation of ions was implemented by a stepwise increase of the source collision voltage (Vc). The value of Vc50 is defined as the cone voltage resulting in 50% dissociation of a complex [35]. The Vc50 values (Figure 3) obtained through ESI-MS were 76.16 V for the EGCG–SOD1 complex, 70.05 V for the ECG–SOD1 complex, and 45.06 V for the EGC–SOD1 complex. The results suggest that the Vc50 value of EGCG is higher than those of ECG and EGC and is in agreement with the above mentioned binding affinity study. Several reports indicated that Vc50 is correlated with the electrostatic and H-bond interactions of proteins with ligands, and hydrophobic interactions are lost during the desolvation process in MS experiments [36,37,38]. Thus, we speculated that EGCG–SOD1 interactions have additional H-bonds in contrast to ECG–SOD1 and EGC–SOD1.
Collision-Induced Unfolding for Exploring the Changes in the Conformation of the EGCG–SOD1 Complex
The conformational changes (generally collapse or unfolding) of proteins in the gas phase can be measured via MS of collision-induced dissociation coupled to IM. CIU enables the assessment of gas-phase stability and comparison of the transitions or intermediates [39, 40]. To test whether EGCG-bound SOD1 resists unfolding under collision activation, we studied the structural stability of SOD1 and EGCG–SOD1 complex through CIU. The target ions were selected in quadrupole, and then activated to unfold under different trap collision energies (20–38 V) in the trap cell of the instrument. Figure 4 shows the results of the CIU experiment performed on the +11 dimer state of the free/single/double ligand-bound forms. As the accelerating voltage increased, the ions had more extended states compared with those under low-energy conditions. When the trap collision energy was low (20 V), the resulting of drift time distributions of SOD1 and EGCG–SOD1 complex showed no distinct conformation difference. At increased collision energy (34 V), a broad drift time distribution was observed, indicating the presence of different unfolded conformations of the protein. As previously reported, the compact conformation, partially unfolding conformation, and unfolding conformation of SOD1 under collision activation were observed [32]. The three conformations were defined A, B, and C, respectively. The content order of C conformation at the trap energy of 34 V is free >single >double bound EGCG-SOD1. Similar results were observed at other collision voltages. These results revealed that the ligand-bound protein has a lower tendency to unfold than free proteins, and two-ligands bound protein is more stable than the one-ligand-bound protein. The influence of the number of EGCGs binding to SOD1 was assessed through CIU fingerprints. The CIU fingerprint of SOD1 is dramatically different from that of the EGCG–SOD1 complex. Figure 5 shows that the conformation of the protein has a greater tendency to shift in low energy than ligand-bound SOD1. These results showed that EGCG can stabilize SOD1 conformation and further retard the dissociation and unfolding process of SOD1.
Investigating the Effect of EGCG on apo-SOD1 Aggregation through ThT Fluorescence Assay
Apo-SOD1 aggregation can be induced by TFE at 5.5 pH [41]. TFE-induced aggregation can be detected by measuring the enhancement of ThT fluorescence after the ThT dye binds to the aggregates. The contributions of EGCG on the prevention of aggregates of TFE-induced apo-SOD1 were investigated by ThT binding assays. We investigated the changes in ThT fluorescence after incubating apo-SOD1 with various folded excess of EGCG. Figure 6 shows the reduction of ThT fluorescence for the apo-SOD1 incubation with EGCG at each time point. The results suggest the content of the aggregates decrease when EGCG binds to apo-SOD1, and thus has a negative relationship with EGCG dose.
Monitoring the Hydrophobic and Conformational Change of apo-SOD1 through ANS and CD Experiments
The denaturant effect of TFE on protein structure interferes with hydrophobic forces that stabilize protein structure [42]. In the presence of TFE, SOD1 mutants expose their hydrophobic faces, resulting in protein aggregation. ANS fluorescence dyes can be used to monitor the change of protein hydrophobic [43, 44], and thus ANS experiment was performed to test whether EGCG can stabilize apo-SOD1 structure and suppress the hydrophobic exposure of apo-SOD1. The control emitted at 520 nm, whereas the sample had a blue shift after ANS attached to the hydrophobic surfaces of the proteins (Figure 7), emitting at around 480 nm. This finding is consistent with a previous report [45]. Meanwhile, the results suggested that protein hydrophobic exposure is reduced in the presence of EGCG. We speculated that EGCG bound with the hydrophobic region prevented the exposure of the hydrophobic face and stabilized the protein structure. The signal change of the negative band centered at 218 nm, which is the characterization of β-sheet, is linked to the changed content of the β-sheets. Protein aggregation increases with increasing number of β-sheet structures [46]. CD analysis (Figure 8) indicated that apo-SOD1 incubated for 6 days has a significantly higher number of β-sheet structures than those incubated with EGCG at 1:10 moral ratio, indicating that EGCG can inhibit apo-SOD1 aggregation.
Conclusions
ESI-IM-MS is suited for the detection of noncovalent interactions and probe solution-phase binding interactions. In this study, ESI-IM-MS exhibits its advantages in the investigation of the interaction between SOD1 and catechins. Vc50 evaluation in the CID experiments provided the stability of the noncovalent complex in the gas phase and relative affinity ranking of catechins as ligands bound to SOD1. We speculated that the differences among the three catechins in terms of their capability to stabilize the SOD1 structure are attributed to the number of their hydroxyl groups, which can form hydrogen bonds in protein–ligand interactions. However, after ligand binding, the enhancement in SOD1 stability was not detected by CID. CIU is introduced to analyze SOD1 stability and its conformational changes, and thus has great potential for the identification of the effects of ligands on SOD1 stability. CIU results revealed that EGCG-bound complex can maintain the compact state of SOD1 than free SOD1 under increased collision voltage in the gas phase. CIU fingerprint experiment is a useful tool for the determination of the differences among the unfolding pathways of proteins and complexes.
Conventional biophysical approach was employed to assess the effectiveness of EGCG as ligand. Changes in hydrophobicity and β-sheet content were emphasized because these properties often affect the protein aggregation. The results of fluorescence and CD experiments showed that EGCG can interact with proteins and inhibit the apo-SOD1 aggregation in vitro. ESI-IM-MS is a powerful tool for the investigation of noncovalent interactions of proteins and ligands and changes in protein conformation.
References
Lacomblez, L., Bensimon, G., Leigh, P N.: Dose-ranging study of riluzole in a myotrophic lateral sclerosis. Lancet 347, 1425–1431(1996)
Silverman, J.M., Fernando, S.M., Grad, L.I., Hill, A.F., Turner, B.J., Yerbury, J.J., Cashman, N.R.: Disease mechanisms in ALS: misfolded SOD1 transferred through exosome-dependent and exosome-independent pathways. Cell. Mol. Neurobiol. 36, 377–381 (2016)
Hayashi, Y., Homma, K., Ichijo, H.: SOD1 in neurotoxicity and its controversial roles in SOD1 mutation-negative ALS. Advances in biological regulation. 60, 95–104 (2016)
Rotunno, M.S., Bosco, D.A.: An emerging role for misfolded wild-type SOD1 in sporadic ALS pathogenesis. Front. Cell. Neurosci. 7, 253 (2013)
Anzai, I., Toichi, K., Tokuda, E., Mukaiyama, A., Akiyama, S., Furukawa, Y.: Screening of drugs inhibiting in vitro oligomerization of Cu/Zn-superoxide dismutase with a mutation causing amyotrophic lateral sclerosis. Front Mol Biosci. 3, 40 (2016)
Cabrera, C., Artacho, R., Giménez, R.: Beneficial effects of green tea—a review. J. Am. Coll. Nutr. 25, 79–99 (2006)
Koh, S.H., Lee, S.M., Kim, H.Y., Lee, K.Y., Lee, Y.J., Kim, H.T., Kim, J., Kim, M.H., Hwang, M.S., Song, C., Yang, K.W., Lee, K.W., Kim, S.H., Kim, O.H.: The effect of epigallocatechin gallate on suppressing disease progression of ALS model mice. Neurosci. Lett. 395, 103–107 (2006)
Bieschke, J., Russ, J., Friedrich, R.P., Ehrnhoefer, D.E., Wobst, H., Neugebauer, K., Wanker, E.E.: EGCG remodels mature α-synuclein and amyloid-β fibrils and reduces cellular toxicity. Proc. Natl. Acad. Sci. 107, 7710–7715 (2010)
Ehrnhoefer, D.E., Bieschke, J., Boeddrich, A., Herbst, M., Masino, L., Lurz, R., Engemann, S., Pastore, A., Wanker, E.E.: EGCG redirects amyloidogenic polypeptides into unstructured, off-pathway oligomers. Nat. Struct. Mol. Biol. 15, 558–566 (2008)
Rambold, A.S., Miesbauer, M., Olschewski, D., Seidel, R., Riemer, C., Smale, L., Brumm, L., Levy, M., Gazit, E., Oesterhelt, D., Baier, M., Becker, C.F., Engelhard, M., Winklhofer, K.F., Tatzelt, J.: Green tea extracts interfere with the stress-protective activity of PrP and the formation of PrP. J. Neurochem. 107, 218–229 (2008)
Ehrnhoefer, D.E., Duennwald, M., Markovic, P., Wacker, J.L., Engemann, S., Roark, M., Legleiter, J., Marsh, J.L., Thompson, L.M., Lindquist, S., Muchowski, P.J., Wanker, E.E.: Green tea (–)-epigallocatechin-gallate modulates early events in huntingtin misfolding and reduces toxicity in Huntington's disease models. Hum. Mol. Genet. 15, 2743–2751 (2006)
Ewald, C., Christen, M.T., Watson, R.P., Mihajlovic, M., Zhou, T., Honegger, A., Pluckthun, A., Caflisch, A., Zerbe, O.: A combined NMR and computational approach to investigate peptide binding to a designed armadillo repeat protein. J. Mol. Biol. 427, 1916–1933 (2015)
Llano-Sotelo, B., Chow, C.S.: RNA-aminoglycoside antibiotic interactions: fluorescence detection of binding and conformational change. Biorg. Med. Chem. Lett. 9, 213–216 (1999)
Gavin, A.C., Maeda, K., Kuhner, S.: Recent advances in charting protein-protein interaction: mass spectrometry-based approaches. Curr. Opin. Biotechnol. 22, 42–49 (2011)
Lanucara, F., Holman, S.W., Gray, C.J., Eyers, C.E.: The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics. Nat. Chem. 6, 281–294 (2014)
Li, S., Jia, J., Gao, X.G., He, X.L., Li, J.P.: Analysis of antibiotics from liquid sample using electrospray ionization-ion mobility spectrometry. Anal. Chim. Acta. 720, 97–103 (2012)
Erba, E.B., Zenobi, R.: Mass spectrometric studies of dissociation constants of noncovalent complexes. Annu. Rep. C Phys. Chem. 107, 199 (2011)
Rogniaux, H., Van Dorsselaer, A., Barth, P., Biellmann, J.F., Barbanton, J., van Zandt, M., Chevrier, B., Howard, E., Mitschler, A., Potier, N., Urzhumtseva, L., Moras, D., Podjarny, A.: Binding of aldose reductase inhibitors: correlation of crystallographic and mass spectrometric studies. J. Am. Soc. Mass Spectrom. 10, 635–647 (1999)
Verbeck, G.F., Ruotolo, B.T., Sawyer, H.A., Gillig, K.J., Russell, D.H.: A fundamental introduction to ion mobility mass spectrometry applied to the analysis of biomolecules. J. Biomol. Techniques: JBT. 13, 56–61 (2002)
Susa, A.C., Wu, C., Bernstein, S.L., Dupuis, N.F., Wang, H., Raleigh, D.P., Shea, J.E., Bowers, M.T.: Defining the molecular basis of amyloid inhibitors: human islet amyloid polypeptide–insulin interactions. J. Am. Chem. Soc. 136, 12912–12919 (2014)
Kloniecki, M., Jablonowska, A., Poznanski, J., Langridge, J., Hughes, C., Campuzano, I., Giles, K., Dadlez, M.: Ion mobility separation coupled with MS detects two structural states of Alzheimer's disease A beta 1-40 peptide oligomers. J. Mol. Biol. 407, 110–124 (2011)
Williams, D.M., Pukala, T.L.: Novel insights into protein misfolding diseases revealed by ion mobility-mass spectrometry. Mass Spectrom. Rev. 32, 169–187 (2013)
Uetrecht, C., Rose, R.J., van Duijn, E., Lorenzen, K., Heck, A.J.: Ion mobility mass spectrometry of proteins and protein assemblies. Chem. Soc. Rev. 39, 1633–1655 (2010)
Eschweiler, J.D., Rabuck-Gibbons, J.N., Tian, Y., Ruotolo, B.T.: CIUSuite: a quantitative analysis package for collision induced unfolding measurements of gas-phase protein ions. Anal. Chem. 87, 11516–11522 (2015)
Zhang, H., Cui, W.D., Gross, M.L.: Mass spectrometry for the biophysical characterization of therapeutic monoclonal antibodies. FEBS Lett. 588, 308–317 (2014)
Han, L.J., Hyung, S.J., Mayers, J.J.S., Ruotolo, B.T.: Bound anions differentially stabilize multiprotein complexes in the absence of bulk solvent. J. Am. Chem. Soc. 133, 11358–11367 (2011)
Han, L.J., Hyung, S.J., Ruotolo, B.T.: Bound cations significantly stabilize the structure of multiprotein complexes in the gas phase. Angew. Chem. Int. Ed. 51, 5692–5695 (2012)
Zhao, Y., Singh, A., Li, L., Linhardt, R.J., Xu, Y., Liu, J., Woods, R.J., Amster, I.J.: Investigating changes in the gas-phase conformation of antithrombin III upon binding of arixtra using traveling wave ion mobility spectrometry (TWIMS). Analyst. 140, 6980–6989 (2015)
Tian, Y., Han, L., Buckner, A.C., Ruotolo, B.T.: Collision induced unfolding of intact antibodies: rapid characterization of disulfide bonding patterns, glycosylation, and structures. Anal. Chem. 87, 11509–11515 (2015)
Yamazaki, Y., Takao, T.: Metalation states versus enzyme activities of Cu, Zn-superoxide dismutase probed by electrospray ionization mass spectrometry. Anal. Chem. 80, 8246–8252 (2008)
Borges-Alvarez, M., Benavente, F., Barbosa, J., Sanz-Nebot, V.: Separation and characterization of superoxide dismutase 1 (SOD-1) from human erythrocytes by capillary electrophoresis time-of-flight mass spectrometry. Electrophoresis. 33, 2561–2569 (2012)
Zhuang, X., Liu, S., Zhang, R., Song, F., Liu, Z., Liu, S.: Identification of unfolding and dissociation pathways of superoxide dismutase in the gas phase by ion-mobility separation and tandem mass spectrometry. Anal. Chem. 86, 11599–11605 (2014)
Mazzitelli, C.L., Chu, Y.J., Reczek, J.J., Iverson, B.L., Brodbelt, J.S.: Screening of threading bis-intercalators binding to duplex DNA by electrospray ionization tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 18, 311–321 (2007)
Wan, C., Cui, M., Song, F., Liu, Z., Liu, S.: A study of the non-covalent interaction between flavonoids and DNA triplexes by electrospray ionization mass spectrometry. Int. J. Mass spectrom. 283, 48–55 (2009)
Potier, N., Donald, L.J., Chernushevich, I., Ayed, A., Ens, W., Arrowsmith, C.H., Standing, K.G., Duckworth, H.W.: Study of a noncovalent trp repressor: DNA operator complex by electrospray ionization time-of-flight mass spectrometry. Protein Sci. 7, 1388–1395 (1998)
El-Kabbani, O., Rogniaux, H., Barth, P., Chung, R.P.T., Fletcher, E.V., Van Dorsselaer, A., Podjarny, A.: Aldose and aldehyde reductases: correlation of molecular modeling and mass spectrometric studies on the binding of inhibitors to the active site. Proteins–Struct. Funct. Genet. 41, 407–414 (2000)
Stojko, J., Fieulaine, S., Petiot-Becard, S., Van Dorsselaer, A., Meinnel, T., Giglione, C., Cianferani, S.: Ion mobility coupled to native mass spectrometry as a relevant tool to investigate extremely small ligand-induced conformational changes. Analyst. 140, 7234–7245 (2015)
Griffey, R.H., Sannes-Lowery, K.A., Drader, J.J., Mohan, V., Swayze, E.E., Hofstadler, S.A.: Characterization of low-affinity complexes between RNA and small molecules using electrospray ionization mass spectrometry. J. Am. Chem. Soc. 122, 9933–9938 (2000)
Mehmood, S., Allison, T.M., Robinson, C.V.: Mass spectrometry of protein complexes: from origins to applications. Annu. Rev. Phys. Chem. 66, 453–474 (2015)
Niu, S., Rabuck, J.N., Ruotolo, B.T.: Ion mobility-mass spectrometry of intact protein-ligand complexes for pharmaceutical drug discovery and development. Curr. Opin. Chem. Biol. 17, 809–817 (2013)
Stathopulos, P.B., Rumfeldt, J.A., Scholz, G.A., Irani, R.A., Frey, H.E., Hallewell, R.A., Lepock, J.R., Meiering, E.M.: Cu/Zn superoxide dismutase mutants associated with amyotrophic lateral sclerosis show enhanced formation of aggregates in vitro. Proc. Natl. Acad. Sci. USA. 100, 7021–7026 (2003)
Muench, C., Bertolotti, A.: Exposure of hydrophobic surfaces initiates aggregation of diverse ALS-causing superoxide dismutase-1 mutants. J. Mol. Biol. 399, 512–525 (2010)
Semisotnov, G.V., Rodionova, N.A., Razgulyaev, O.I., Uversky, V.N., Gripas, A.F., Gilmanshin, R.I.: study of the molten globule intermediate state in protein folding by a hydrophobic fluorescent-probe. Biopolymers. 31, 119–128 (1991)
Mulligan, V.K., Kerman, A., Laister, R.C., Sharda, P.R., Arslan, P.E., Chakrabartty, A.: Early steps in oxidation-induced SOD1 misfolding: implications for non-amyloid protein aggregation in familial ALS. J. Mol. Biol. 421, 631–652 (2012)
Tiwari, A., Liba, A., Sohn, S.H., Seetharaman, S.V., Bilsel, O., Matthews, C.R., Hart, P.J., Valentine, J.S., Hayward, L.J.: Metal deficiency increases aberrant hydrophobicity of mutant superoxide dismutases that cause amyotrophic lateral sclerosis. J. Biol. Chem. 284, 27746–27758 (2009)
Leal, S.S., Cardoso, I., Valentine, J.S., Gomes, C.M.: Calcium ions promote superoxide dismutase 1 (SOD1) aggregation into non-fibrillar amyloid a link to toxic effects of calcium overload in amyotrophic lateral sclerosis (ALS)? J. Biol. Chem. 288, 25219–25228 (2013)
Acknowledgments
This work was supported by the National Natural Science Foundation of China (no. 21673219 and 81530094).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing financial interest.
Electronic supplementary material
ESM 1
(DOCX 924 kb)
Rights and permissions
About this article
Cite this article
Zhao, B., Zhuang, X., Pi, Z. et al. Determining the Effect of Catechins on SOD1 Conformation and Aggregation by Ion Mobility Mass Spectrometry Combined with Optical Spectroscopy. J. Am. Soc. Mass Spectrom. 29, 734–741 (2018). https://doi.org/10.1007/s13361-017-1864-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1864-z