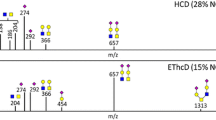
Abstract
Tris(hydroxymethyl)aminomethane (Tris) is one of the most frequently used buffer ingredients. Among other things, it is recommended and is usually used for lectin-based affinity enrichment of glycopeptides. Here we report that sialic acid, a common ‘capping’ unit in both N- and O-linked glycans may react with this chemical, and this side reaction may compromise glycopeptide identification when ETD spectra are the only MS/MS data used in the database search. We show that the modification may alter N- as well as O-linked glycans, the Tris-derivative is still prone to fragmentation both in ‘beam-type’ CID (HCD) and ETD experiments, at the same time—since the acidic carboxyl group was ‘neutralized’—it will display a different retention time than its unmodified counterpart. We also suggest solutions that—when incorporated into existing search engines—may significantly improve the reliability of glycopeptide assignments.

ᅟ





Similar content being viewed by others
References
Varki, A., Cummings, R.D., Esko, J.D., Freeze, H.H., Stanley, P., Bertozzi, C.R., Hart, G.W., Etzler, M.E. (eds.): Essentials of Glycobiology, 2nd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor (2009)
Medzihradszky, K.F.: Characterization of protein N-glycosylation. Methods Enzymol. 405, 116–138 (2005)
Peter-Katalinić, J.: Methods in enzymology: O-glycosylation of proteins. Methods Enzymol. 405, 139–171 (2005)
Dodds, E.D.: Gas-phase dissociation of glycosylated peptide ions. Mass Spectrom. Rev. 31, 666–682 (2005)
Settineri, C.A., Medzihradszky, K.F., Masiarz, F.R., Burlingame, A.L., Chu, C., George-Nascimento, C.: Characterization of O-glycosylation sites in recombinant B-chain of platelet-derived growth factor expressed in yeast using liquid secondary ion mass spectrometry, tandem mass spectrometry and Edman sequence analysis. Biomed. Environ. Mass Spectrom. 19, 665–676 (1990)
Medzihradszky, K.F., Besman, M.J., Burlingame, A.L.: Structural characterization of site-specific N-glycosylation of recombinant human factor VIII by reversed-phase high-performance liquid chromatography-electrospray ionization mass spectrometry. Anal. Chem. 69, 3986–3994 (1997)
Stimson, E., Hope, J., Chong, A., Burlingame, A.L.: Site-specific characterization of the N-linked glycans of murine prion protein by high-performance liquid chromatography/electrospray mass spectrometry and exoglycosidase digestions. Biochemistry 38, 4885–4895 (1999)
Schmitt, S., Glebe, D., Alving, K., Tolle, T.K., Linder, M., Geyer, H., Linder, D., Peter-Katalinic, J., Gerlich, W.H., Geyer, R.: Analysis of the pre-S2 N- and O-linked glycans of the M surface protein from human hepatitis B virus. J. Biol. Chem. 274, 11945–11957 (1999)
Hofsteenge, J., Huwiler, K.G., Macek, B., Hess, D., Lawler, J., Mosher, D.F., Peter-Katalinic, J.: C-mannosylation and O-fucosylation of the thrombospondin type 1 module. J. Biol. Chem. 276, 6485–6498 (2001)
Zubarev, R.A., Horn, D.M., Fridriksson, E.K., Kelleher, N.L., Kruger, N.A., Lewis, M.A., Carpenter, B.K., McLafferty, F.W.: Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem. 72, 563–573 (2000)
Syka, J.E.P., Coon, J.J., Schroeder, M.J., Shabanowitz, J., Hunt, D.F.: Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 101, 9528–9533 (2004)
Darula, Z., Medzihradszky, K.F.: Affinity enrichment and characterization of mucin core–1 type glycopeptides from bovine serum. Mol. Cell Proteomics 8, 2515–2526 (2009)
Steentoft, C., Vakhrushev, S.Y., Vester-Christensen, M.B., Schjoldager, K.T., Kong, Y., Bennett, E.P., Mandel, U., Wandall, H., Levery, S.B., Clausen, H.: Mining the O-glycoproteome using zinc-finger nuclease-glycoengineered SimpleCell lines. Nat. Methods 8, 977–982 (2011)
Halim, A., Nilsson, J., Rüetschi, U., Hesse, C., Larson, G.: Human urinary glycoproteomics; attachment site specific analysis of N- and O-linked glycosylations by CID and ECD. Mol. Cell Proteomics 11(4), M111.013649 (2012)
Trinidad, J.C., Barkan, D.T., Gulledge, B.F., Thalhammer, A., Sali, A., Schoepfer, R., Burlingame, A.L.: Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell Proteomics 11, 215–229 (2012)
Yin, X., Bern, M., Xing, Q., Ho, J., Viner, R., Mayr, M.: Glycoproteomics analysis of the secretome of human endothelial cells. Mol. Cell Proteomics 12, 956–978 (2013)
Mikesh, L.M., Ueberheide, B., Chi, A., Coon, J.J., Syka, J.E., Shabanowitz, J., Hunt, D.F.: The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta 1764, 1811–1822 (2006)
Saba, J., Dutta, S., Hemenway, E., Viner, R.: Increasing the productivity of glycopeptides analysis by using higher-energy collision dissociation-accurate mass-product-dependent electron transfer dissociation. Int. J. Proteomics 2012, 560391 (2012)
Bern, M., Kil, Y.J., Becker, C.: Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics. Chapter 13:Unit13.20 (2012) doi:10.1002/0471250953.bil320s40
Chalkley, R.J., Baker, P.R., Medzihradszky, K.F., Lynn, A.J., Burlingame, A.L.: In-depth analysis of tandem mass spectrometry data from disparate instrument types. Mol. Cell Proteomics 7, 2386–2398 (2008)
Sato, C., Kim, J.H., Abe, Y., Saito, K., Yokoyama, S., Kohda, D.: Characterization of the N-oligosaccharides attached to the atypical Asn-X-Cys sequence of recombinant human epidermal growth factor receptor. J. Biochem. 127, 65–72 (2000)
Trinidad, J.C., Schoepfer, R., Burlingame, A.L., Medzihradszky, K.F.: N- and O-glycosylation in the murine synaptosome. Mol. Cell Proteomics 12, 3474–3488 (2013)
Moloney, D.J., Lin, A.I., Haltiwanger, R.S.: The O-linked fucose glycosylation pathway. evidence for protein-specific elongation of O-linked fucose in Chinese hamster ovary cells. J. Biol. Chem. 272, 19046–19050 (1997)
Rossez, Y., Maes, E., Lefebvre Darroman, T., Gosset, P., Ecobichon, C., Joncquel Chevalier Curt, M., Boneca, I.G., Michalski, J.C., Robbe-Masselot, C.: Almost all human gastric mucin O-glycans harbor blood group A; B or H antigens and are potential binding sites for Helicobacter pylori. Glycobiology 22, 1193–1206 (2012)
Darula, Z., Chalkley, R.J., Lynn, A., Baker, P.R., Medzihradszky, K.F.: Improved identification of O-linked glycopeptides from ETD data with optimized scoring for different charge states and cleavage specificities. Amino Acids 41, 321–328 (2011)
Zauner, G., Kozak, R.P., Gardner, R.A., Fernandes, D.L., Deelder, A.M., Wuhrer, M.: Protein O-glycosylation analysis. Biol. Chem. 393, 687–708 (2012)
Pan, G.G., Melton, L.D.: Lactones of disialyl lactose: characterisation by NMR and mass spectra. Carbohydr. Res. 341, 730–737 (2006)
Bassi, R., Riboni, L., Sonnino, S., Tettamanti, G.: Lactonization of GD1b ganglioside under acidic conditions. Carbohydr. Res. 193(C), 141–146 (1989)
Pudelko, M., Lindgren, A., Tengel, T., Reis, C.A., Elofsson, M., Kihlberg, J.: Formation of lactones from sialylated MUC1 glycopeptides. Org. Biomol. Chem. 4, 713–720 (2006)
Nielsen, M.L., Savitski, M.M., Zubarev, R.A.: Improving protein identification using complementary fragmentation techniques in fourier transform mass spectrometry. Mol. Cell Proteomics 4, 835–845 (2005)
Hansen, T.A., Sylvester, M., Jensen, O.N., Kjeldsen, F.: Automated and high confidence protein phosphorylation site localization using complementary collision-activated dissociation and electron transfer dissociation tandem mass spectrometry. Anal. Chem. 84, 9694–9699 (2012)
Darula, Z., Chalkley, R.J., Baker, P., Burlingame, A.L., Medzihradszky, K.F.: Mass spectrometric analysis; automated identification and complete annotation of O-linked glycopeptides. Eur. J. Mass Spectrom. 16, 421–428 (2010)
Hägglund, P., Bunkenborg, J., Elortza, F., Jensen, O.N., Roepstorff, P.: A new strategy for identification of N-glycosylated proteins and unambiguous assignment of their glycosylation sites using HILIC enrichment and partial deglycosylation. J. Proteome Res. 3, 556–566 (2004)
Froesch, M., Bindila, L., Zamfir, A., Peter-Katalinić, J.: Sialylation analysis of O-glycosylated sialylated peptides from urine of patients suffering from Schindler’s disease by Fourier transform ion cyclotron resonance mass spectrometry and sustained off-resonance irradiation collision-induced dissociation. Rapid Commun. Mass Spectrom. 17, 2822–2832 (2003)
Parker, B.L., Thaysen-Andersen, M., Solis, N., Scott, N.E., Larsen, M.R., Graham, M.E., Packer, N.H., Cordwell, S.J.: Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J. Proteome Res. 12, 5791–5800 (2013)
Acknowledgments
The authors gratefully acknowledge the contribution of Ralf Schoepfer and Jonathan Trinidad in the mouse synaptosome study, and thank Marshall Bern for performing the Byonic database search, Adam Kerenyi for writing the script used for HCD peak list filtering, and Zoltan Kupihar for useful discussions and technical assistance.
K.F.M. was supported by NIH grant NIGMS 8P41GM103481, and by the Howard Hughes Medical Institute (to the Bio-Organic Biomedical Mass Spectrometry Resource at UCSF, Director: A.L. Burlingame), and by the following grants: OTKA 105611 (to Z.D.), and BAROSS-DA07-DA-ESZK-07-2008-0036 (to the Biological Research Centre, HAS, director: P. Ormos). Z.D. was supported by the Janos Bolyai Fellowship of the HAS.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Darula, Z., Medzihradszky, K.F. Glycan Side Reaction May Compromise ETD-Based Glycopeptide Identification. J. Am. Soc. Mass Spectrom. 25, 977–987 (2014). https://doi.org/10.1007/s13361-014-0852-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-014-0852-9