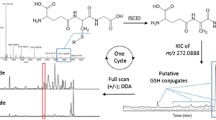
Abstract
Drugs can be metabolically activated to soft and hard electrophiles, which are readily trapped by glutathione (GSH) and cyanide (CN), respectively. These adducts are often detected and structurally characterized using separate tandem mass spectrometry methods. We describe a new method for simultaneous screening of GSH and CN adducts using precursor ion (PI) and neutral loss (NL) scans-dependent product ion spectral acquisition and data mining tools on an triple quadrupole linear ion trap mass spectrometry. GSH, potassium cyanide, and their stable isotope labeled analogues were incubated with liver microsomes and a test compound. Negative PI scan of m/z 272 for detection of GSH adducts and positive NL scans of 27 and 29 Da for detection of CN adducts were conducted as survey scans to trigger acquisition of enhanced resolution (ER) spectrum and subsequent enhanced product ion (EPI) spectrum. Post-acquisition data mining of EPI data set using NL filters of 129 and 27 Da was then performed to reveal the GSH adducts and CN adducts, respectively. Isotope patterns and EPI spectra of the detected adducts were utilized for identification of their molecular weights and structures. The effectiveness of this method was evaluated by analyzing reactive metabolites of nefazodone formed from rat liver microsomes. In addition to known GSH- and CN-trapped reactive metabolites, several new CN adducts of nefazodone were identified. The results suggested that current approach is highly effective in the analysis of both soft and hard reactive metabolites and can be used as a high-throughput method in drug discovery.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Xenobiotics, including drugs and environmental toxicants, can be metabolically activated by cytochrome P450 enzyme (CYPs) or other enzymes to form reactive intermediates that can covalently bind to cellular components such as proteins and nucleic acids. According to the principle of hard and soft acid base (HSAB), reactive intermediates can be classified into “soft” and “hard” species [1]. The soft reactive intermediates contain functional groups of large radius that are easy to polarize. In comparison, the hard reactive metabolites usually have a small radius and are more difficult to polarize. The majority of reactive metabolites can be categorized as soft electrophiles, including quinones, quinone-imines, iminoquinone methides, epoxides, arene oxides, and nitrenium ions, which can readily react with soft nucleophiles such as the sulfhydryl group in glutathione (GSH) and cysteine residue in proteins. The hard reactive metabolites, most often aldehydes and iminium species, are readily reactive to hard nucleophiles such as bases in DNA and lysine residues in proteins [2, 3]. Direct detection of the reactive intermediates has proven to be almost impossible due to their transient nature. Instead, the most commonly utilized approach is to capture the reactive intermediate with a trapping agent, leading to formation of stable adducts of reactive metabolites that can be detected and characterized using LC-MS/MS techniques [4–6]. These methods have been extensively applied to efforts in the minimization of bioactivation for lead compounds at the drug discovery stage [7–11].
Commonly used trapping agents for soft electrophiles are GSH and other thiol-based electrophiles such as N-acetyl-cysteine and cysteine [12]. A number of derivatives of GSH such as GSH ethyl ester, dansyl-GSH, brominated GSH, or quaternary ammonium GSH have also been utilized to facilitate MS detection or quantitation [13–16]. Introduction of stable isotope-labeled GSH analogues such as 13C2 15N-GSH into an incubation medium provides GSH-trapped reactive metabolites with unique isotope patterns that can be used either in isotope pattern-dependent scan or post-acquisition data mining [17–20]. Hard reactive intermediates cannot be efficiently trapped by GSH or its derivatives. Alternatively, potassium cyanide (KCN) has been proven to be a useful agent for trapping compounds that form hard reactive intermediate such as iminium ion. Other trapping agents, including semicarbazide, methoxylamine, histidine, and lysine can be used to trap other hard electrophiles such as aldehydes [21]. In addition, radiolabeled GSH and CN have been used as trapping agents to generate radio-labeled adducts, which can be measured by radioactivity detection to quantitatively evaluate the bioactivation potentials of drug candidates [22, 23].
A number of LC/MS methodologies have been developed and applied for detection and structural characterization of GSH- and CN-trapped reactive metabolites formed in incubations. These LC/MS approaches can be divided into two categories: triple quadrupole MS- and high resolution MS (HR-MS)-based methodologies. Detection of GSH and CN adducts by triple quadrupole or triple quadrupole-linear ion trap (Qtrap) mass spectrometry is based on their unique fragmentation patterns. For example, neutral loss (NL) of 129 Da in the positive ion mode that corresponds to loss of pyroglutamic acid is the most commonly used scanning function for detection of GSH adducts by triple quadrupole instruments [18, 24]. Alternatively, GSH adducts produce characteristic product ion at m/z 272 in negative mode, which can be used in precursor ion (PI) scan for detection [25]. Qtrap has been shown to be more efficient in screening of GSH adducts than triple quadrupole instruments. In Qtrap analysis, either positive NL scan of 129 Da or negative PI scan of m/z 272 can serve as the survey scan to trigger the acquisition of positive product ion spectra of the detected adducts [13, 26]. In addition, predictive multiple reaction monitoring (MRM) based on predicted fragmentations of GSH adducts were used for more sensitive detection of GSH adducts by Qtrap [26]. In a similar manner, cyanide adducts can be detected using NL scan of 27 Da, a characteristic fragmentation pathway of CN adducts, followed by acquisition of product ion spectra for structural elucidation on a triple quadrupole instrument. Argoti et al. developed a relatively high throughput screening method for the detection of the iminium ion from 14 alicyclic amines by NL scanning of 27 and 29 Da using KCN and K13C15N as trapping agents [27]. The inclusion of stable isotope labeled KCN provided unique isotope pattern to facilitate the elimination of false positives. In contrast, detection of GSH and CN adducts of reactive metabolites by HR-MS relays on characteristics of protonated molecules of the adducts, including their predictable ranges of mass defects or unique isotope patterns. In addition, background subtraction processing that extracts ion species present in accurate full MS spectra of a test samples but not in a control samples is applied for detection of GSH adducts in vitro and in animals [15, 20, 28]. Recently, Barbara et al. reported a LC-time-of-flight (TOF) MSE post-acquisition data mining method for reactive metabolite screening. Post-acquisition mining using mass defect filter (MDF) that detect drug metabolites based on predicable ranges of their mass defect values, automated comparison of sample and control, and time-alignment of high- and low-energy MSE data enabled detection of reactive metabolites without the need for stable- or radio-isotope-labeled trapping agents [29].
Traditionally, in vitro screening of soft and hard reactive metabolites is carried out using separate GSH and CN trapping experiments, each of which employs a specific LC/MS method. To increase the productivity of reactive metabolite screening, significant efforts have been made to combine the two experiments. The task can be readily accomplished with HR-MS-based approached. Rousu et al. reported a screening method utilizing stable isotope labeled GSH, KCN, and semicarbazide as trapping agents and TOF MS as analytical tool [30]. MDF processing of high resolution full MS data set provided unambiguous identification of both hard and soft reactive metabolites. A different approach was presented by Lim et al. in their work of minimizing bioactivation of a drug candidate, in which S9 incubation in the presence of GSH and CN, along with their stable isotope analogues, was subject to HR-MS and isotope pattern dependent MSn acquisition on Orbitrap MS [11]. Mitchell et al. used a peptide consisting of eleven amino acids including a terminus GSH and additional nucleophilic residues (e.g., lysine) for simultaneously trapping of both soft and hard electrophiles [31]. The assay utilized a surface-enhanced laser detection/ionization (SELDI)-TOF MS system without chromatography separation. Drug-peptide adducts were observed as peaks with higher mass relative to the trapping peptide in the spectra. In contrast, simultaneous detection of both GSH and CN adducts by a traditional triple quadruple or Qtrap instrument is very challenging. The NL scan and PI scan that are used to detect GSH and CN adducts have relatively slow speeds so that it is not practical to include multiple NL and PI scans for simultaneous detection of GSH and CN adducts in a single analysis, not to mention to accommodate a polarity switch. Alternatively, Yan et al. developed a method for simultaneous capturing and detection of both soft and hard reactive intermediate by using a specially designed trapping agent γ-glutamylcysteinlysine (γ-GSK) [32]. In γ-GSK, a lysine residue replaced the glycine. The amine group of the lysine can react with hard electrophiles, while the sulfhydryl moiety can be reactive to the soft electrophiles. Since the pyroglutamic acid portion remains unchanged, both hard and soft adducts can be detected using NL scan of 129 Da. However, the effectiveness of this method has not been evaluated for certain classes of GSH adducts that do not fragment by NL of 129 Da [27, 33]. Recently, updated triple quadrupole linear ion trap instrument (API Qtrap 5500) has been introduced, which offers improved scan speed, sensitivity, and polarity switch efficiency. The hybrid Qtrap instrument is often used as a triple quadrupole instrument for drug and metabolite quantification in drug metabolism and pharmacokinetics studies. On the other hand, its unique scanning functions, including PI scan, NL scan, predictive MRM, and multiple ion monitoring that can trigger MS/MS acquisition, serve well for profiling and identification of drug metabolites and reactive intermediates [34]. There is a new trend that uses a single LC/MS platform in drug metabolism and pharmacokinetics studies in drug discovery, which can significantly reduce cost and increase productivity. Examples include drug quantification in a metabolic stability experiment, followed by metabolic soft-spot identification, and identification of plasma metabolites in humans and preclinical species, followed by quantification of major human metabolites. HR-MS and Qtrap instruments are the mass spectrometers of choice for these applications. The major goal of the current study was to develop a practical method for simultaneous detection of GSH and CN adducts generated in the same incubation by taking advantage of high scan speed and fast polarity switch on new Qtrap 5500. Nefazodone, a drug that can be bioactivated to generate both soft and hard reactive intermediates, was used as a model compound to demonstrate the effectiveness of the current approach.
2 Experimental
2.1 Materials
Pooled rat liver microsomes containing 20 mg protein/mL were obtained from BD Biosciences (Woburn, MA, USA). NADPH, GSH, KCN, K13C15N, and nefazodone were purchased from Sigma-Adrich (St. Louis, MO, USA). The Oasis HLB 3 mL cartridge was from Waters (Milford, MA, USA). 13C 152 N-GSH was purchased from Cambridge isotope laboratories (Andover, MA, USA)
2.2 Microsomal Incubation and Sample Preparation
The incubation mixture of 2 mL in volume consisted of the following components: 1 mg/mL of rat liver microsomal protein, 1 mM NADPH, 1 mM GSH/13C 152 N-GSH (1:1, 0.5 mM each), and 1 mM KCN/K13C15N (1:1,0.5 mM each), and 30 μM nefazodone as substrate. The biotransformation reactions were initiated by the addition of the NADPH solution after a 3 min preincubation, and were stopped by a solid phase extraction. The incubation time was 30 min. Control incubations were carried out in the same fashion without the addition of NADPH. The Oasis HLB 3 mL cartridge was used for solid phase extraction. Before loading the sample, the cartridge was washed with 1 mL of methanol and equilibrated with 1 mL of water. Then, 2 mL of sample was loaded and washed with 1 mL of water for two times. The sample was eluted out with 2 mL of methanol. The sample was dried down under nitrogen gas and reconstituted with 150 μL of 50% methanol in water (vol:vol). An aliquot of 10 μL was analyzed by LC-MS.
2.3 Instrumentation
The LC-MS analysis was conducted on Shimadzu LC-20 AD HPLC pumps and Leap CTC PAL autosampler coupled with Qtrap 5500 (AB SCIEX, Foster City, CA, USA). The separation was carried on an Agilent Eclipse plus C18 column (2.1 × 100 mm, 1.8 μm). Mobile phase A was 0.1% formic acid in water and mobile phase B was 0.1% formic acid in acetonitrile. The gradient was: 10% B at 0 min, 10% B at 2 min, 70% B at 12 min, 90% B at 14 min, 90% B at 18 min, 10% B at 18.2 min, stopped at 20 min. Flow rate was 0.35 mL/min. The Qtrap 5500 was operated in Turbospray mode and the operating parameters were optimized and set at the following values: curtain gas 30 psi, GAS1 50 psi, GAS2 50 psi, source temperature 500 °C, CAD gas high, ionspray voltage 4500 V for positive mode and –4500 V for negative mode, declustering potential 100 V for positive mode and –100 V for negative mode, entrance potential 15 V in positive mode and –15 V in negative mode, and collision cell exit potential 15 V in positive mode and –15 V in negative mode. Negative PI scan (m/z 272 for GSH adducts) and positive NL scan (27 and 29 Da for CN adducts) were utilized as survey scans to trigger the acquisition of positive ER and EPI spectra. Criteria for the information-dependent acquisition (IDA) of ER and EPI were set for most intensive peak, which exhibited counts higher than 1000 cps. The scan speed for PI, NL, ER, and EPI were 2000 Da/s, 2000 u/s, 250 u/s, and 20,000 u/s, respectively. The settling time for polarity switch was 100 ms. Collision energy was set at –35 eV for negative PI scan, 20 eV for positive NL scan, and 50 eV with 20 eV of collision energy spread (CES) for positive EPI scans. The total cycle time was 2.45 s. Dynamic fill time was used to ensure the optimal enhanced product ion scan quality at different concentrations.
2.4 Data Mining
The LC-MS data acquired on Qtrap were processed using Analyst 1.5.1. The EPI data set was filtered using the script of IDA tracer extractor. NL filters with a range of 128.5 to 129.5 Da and a range of 26.5 to 27.5 Da were applied to the data to generate extracted chromatograms of GSH adducts and CN adducts, respectively. Identity of each detected adduct was confirmed by inspecting the ER spectrum of the molecular ion for expected isotope patterns. The product ion spectrum of each detected adduct was further recovered from the same EPI data set for elucidation of their structures.
3 Result and Discussion
3.1 Analytical Strategy
The analytical strategy used in this study is depicted in Figure 1. In order to trap both soft and hard reactive metabolites, GSH and KCN were included in the same experiment. Stable isotope labeled GSH and KCN were also added at equal amounts as the non-labeled versions to facilitate the confirmation of the detected adducts of reactive metabolites during data mining process. Negative PI scan of m/z 272 for detection of GSH adducts and positive NL scans of 27 and 29 Da for detection of CN adducts were conducted as survey scans. Negative PI scan of m/z 272 has been shown to provide significantly improved selectivity and sensitivity in detection of unknown GSH adducts [13, 25]. NL scan of 27 Da in positive mode is characteristic of CN adducts but may not be selective enough due to its low molecular mass. Therefore, a NL scan of 29 Da was also included to detect adducts generated from K13C15N. Once the ion intensity of PI or NL scan reached a threshold of 1000 cps, a positive ER scan was conducted to acquire high quality full scan mass spectrum of a narrow mass range around the most abundant ions. The threshold of 1000 cps was selected based on the observed background noise level and desired sensitivity of product ion scan. It was considered that peaks lower than 1000 cps would either be nonspecifically generated from background noise or give spectra of low sensitivity and poor quality in subsequent product ion scan. The monoisotopic peak confirmed in ER scan was then used as a precursor ion in subsequent EPI scan to obtain positive product ion spectra of GSH and CN adducts detected by either PI/ER with polarity switch or NL/ER, respectively.
The recorded EPI data set was further processed for detection of GSH and CN adducts and for elimination of false positive ions present in total ion chromatogram (TIC) of EPI. NL filtering of 129 Da and 27 Da was employed for extraction of GSH and CN adducts from the EPI data set, respectively. In addition, the isotopic pattern of the ER spectrum for each detected peak was also closely inspected to confirm the identification and to exclude any false positive. The isotopic doublets derived from GSH and 13C 152 N-GSH should differ by three mass unit and show a peak intensity ratio around 1:1. The doublets from CN and 13C15N differing by two mass unit should exhibit a ratio of approximately 1:1.3 due to the contribution of 37Cl-containing CN adducts to the 13C15N adduct. Once GSH and CN adducts were detected and confirmed via data processing, their product ion spectra were readily recovered from the EPI data set and used for structural characterization (Figure 1).
The current analytical strategy takes advantages of the high acquisition speed and fast polarity switch on the new API Qtrap 5500 instrument to enable all the data acquisition to be conducted in a single run. Furthermore, it has a polarity switch settling time of only 50 ms that enables the simultaneous acquisition in both positive and negative modes without sacrificing the acquisition time. In the current work, four types of scans, PI, NL, ER, and EPI, and one polarity switch were completed within a cycle time of 2.45 s, fast enough to allow enough points to be collected for each chromatographic peak. In comparison, PI or NL scan on an earlier version of triple quadrupole-linear ion trap instrument (Qtrap 4000) is relatively slow. Therefore, combined NL and PI scans cannot be accommodated in a single run. In addition, the polarity switch on a Qtrap 4000 instrument would require a settling time of 700 ms, which extends the cycle time significantly.
3.2 Detection and Characterization of Soft Reactive Metabolites
Nefazodone has been demonstrated to undergo bioactivation at the 3-chlorophenylpiperazine motif to form reactive quinone-imine species, which can be trapped by GSH to form adducts [29, 35, 36]. In addition, Alfa-carbon oxidation of the piperazine ring in nefazodone results in iminium ions, which are hard electrophiles and can be trapped by cyanide to form CN adducts [27, 36] (Scheme 1). In this study, nefazodone was chosen as a model compound to examine the effectiveness of the current approach (Figure 1). The TIC of EPI, which was triggered by negative PI scan of m/z 272 for GSH adducts, as well as positive NL scan of 27 and 29 Da for CN adducts, revealed five major adducts, including one GSH adduct (GM5) and four CN adducts (CM4, CM8, CM10, and CM12, Table 1), along with several false positive peaks (Figure 2a). However, many minor GSH and CN adducts of nefazodone were not visible in the TIC due to the high background noise or interference of the false positive signals. NL filtering of 129 Da from the EPI data was able to remove most interference ions and background noises, leading to a significantly improved GSH adduct profile, in which additional five minor GSH adducts (GM1-GM4, and GM6, Table 1) were clearly displayed with few minor interference peaks (Figure 2b). The NL filtering of EPI data also provided better selectivity than direct negative PI scanning of m/z 272 (Figure 2c), in which more false peaks were present. The improved sensitivity of GSH adduct detection of the current analytical strategy may be contributed by the overall increased sensitivity of Qtrap 5500 and the processing of EPI data set with neutral loss filtering. In addition to NL of 129 Da, other unique fragmentation pathways of GSH adducts such as NL of 307 Da or formation of product ion at m/z 149 can be applied as filters to process the EPI data file, which may provide better selectivity and sensitivity than NL filtering of 129 Da in finding certain types of GSH adducts [27, 33].
LC-MS/MS analysis of GSH and CN adducts generated from nefazodone in a rat liver microsome incubation using PI- and NL-dependent EPI scans followed by data mining. (A) TIC of positive EPI triggered by negative PI at m/z 272 and positive NL of 27 Da and 29 Da. (B) Processed EPI profile by NL filter of 129 Da for detection of GSH adducts. (C) TIC of negative PI at m/z 272. (D) Processed EPI profile by NL filter of 27 Da for detection of CN adducts
GM1 and GM2 eluted at 6.3 and 6.5 min, respectively, and showed the identical [M+H]+ ion (Figure 3a) and EPI spectrum (Figure 3b). The [M+H]+ ion at m/z 807 corresponds to the addition of GSH to a dihydroxylated nefazodone metabolite [29, 36]. The ER scan spectrum around m/z 807 exhibited an isotope envelop that is consistent with the presence of both GSH and 13C 152 N-GSH adducts (Figure 3a). In comparison, the ER spectrum of a false positive peak in the profile processed with the NL filter of 129 Da (Figure 2b, labeled with an asterisk (*) at retention time of 9.2 min), did not exhibit the unique isotope pattern (Figure 3c). The EPI spectrum of GM2 displayed diagnostic fragment ions at m/z 678, 534, and 290 (Figure 3b), which is consistent with the product spectrum of a previously reported GSH adduct of dihydroxylated nefazodone generated from human liver microsome incubation [36]. The structure and fragment assignment of the GSH adduct is displayed in Figure 3a. The fragment ion at m/z 290 (addition of 16 Da to the fragment ion at m/z 274 of nefazodone) supported the presence of monohydroxylation, possibly on the 5-ethyl side chain of the triazolone moiety of the molecule. The other monohydroxylation occurred on the 3-chlorophenylpiperazine ring as a para-hydroxylation. The regiochemical assignments for the hydroxylations are based on the previously characterized dihydroxylated metabolites [37, 38]. It was reported that the formation of hydroxylated metabolites on the 5-ethyl side chain and the 3-chlorophenypiperazine were major metabolic pathways of nefazodone in human liver microsomes. Most likely, GM1 and GM2 were formed via the dihydroxylation, followed by formation of the quinone-imine reactive metabolite and the GSH addition to the different positions on the para-hydroxynefazodone (Scheme 1). Detection and identification of the less abundant GM1 indicated that the current methodology provided good sensitivity for minor metabolites.
(a) Positive ER spectrum and proposed structure of nefazodone GSH adduct GM2. (b) Positive EPI spectrum of GM2. (c) Positive ER spectrum of a false positive peak in the EPI processed by NL filter of 129 Da for detection of GSH adducts (Figure 2b, labeled with an asterisk (*) at retention time of 9.2 min)
GM5 was shown as a major peak (7.0 min) in the NL filtered EPI chromatogram (Figure 2b). It shared the identical [M+H]+ ion (Supplementary Information, Figure S1A) and EPI spectrum (Supplementary Information, Figure S1B) as GM3 that was eluted as a small peak at 6.8 min. The [M+H]+ ion at m/z 791 corresponded to addition of GSH to a monohydroxylated nefazodone metabolite. The ER scan spectrum around m/z 791 exhibited an isotope envelop that is consistent with the presence of both GSH and 13C 152 N-GSH adducts (Supplementary Information Figure, S1A). The diagnostic fragment ions at m/z 662, 518, and 274 (Supplementary Information, Figure S1B) are consistent with the previously reported para-hydroxynefazodone GSH adduct that was formed via reactive chlorophenyl quinone-imine ion after monohydroxylation on the 3-chlorophenypiperazine (Scheme 1) [35, 36]. Detection of a pair of isomers is consistent with the observation made by Barbara et al. in their work of detecting GSH adducts using MSE and data-mining [29].
GM4 and GM6 were another pair of minor GSH adducts that shared the identical [M+H]+ and EPI spectrum (Figure 4a and b). They were eluted as minor peaks at retention time of 6.9 and 7.1 min, respectively. The [M+H]+ at m/z of 805 was two mass unit lower than the GSH adducts of dihydroxylated nefazodone. The diagnostic fragment ion at m/z 288 suggested further oxidation of the hydroxyl group on the 5-ethyl side chain of triazolone moiety, resulting in the formation of the corresponding ketone (Figure 4). Same oxidation reaction has been shown to take place on phase I nefazodone metabolites [37, 38]. Therefore, GM4 and GM6 were tentatively designated as a pair of GSH adducts of mono-dehydrogenated dihydroxylation nefazodone (Scheme 1), which is consistent with what previously reported by Barbara et al. [29]. However, in their work, few useful fragments were observed in the product ion spectra for one of the isomers, so that structural elucidation could not be performed. In comparison, enhanced product ion scan in current work enabled the acquisition of high quality product ion spectra for both of the isomers.
3.3 Detection and Characterization of Hard Reactive Metabolites
In the current study, the NL scan of 27 Da for CN adducts and NL scan of 29 Da for 13C15N adducts resulted in similar ion chromatograms in which multiple CN adduct peaks and relative high background noise were observed (data not shown). The TIC of EPI revealed four major CN adducts of nefazodone, while the less abundant ones were not clearly visible (Figure 2a). The NL filter of 27 Da was further applied to the EPI data set to reveal potential CN adducts (Figure 1). As shown in Figure 2d, the filtered EPI data was able to reveal 14 CN adducts with only about five to six minor false positive peaks, which may be derived from the components in biological matrix. NL filter of 29 Da was also applied to the EPI data to reveal the 13C15N adducts, which were identical to those in the profile from NL filtering of 27 Da (data not shown), indicating that NL filtering of 27 Da alone was specific enough to detect the CN adducts. In addition, the isotope pattern of each peak displayed in Figure 2d was inspected via ER data set (Figure 1), further confirming the identity of the CN adduct peaks and the false positive ions. The CN adducts were designated as CM1-CM14 (Figure 2d and Table 1) and their proposed structures and MS/MS fragment pathways are displayed in Scheme 1. All of these adducts were formed via iminium ions, reactive intermediates on the piperazine ring.
CM12 was one of the major CN adducts. Its [M+H]+ at m/z 495 indicated direct conjugation of CN with nefazodone. NL of 27 Da led to formation of the fragment ion at m/z 468 that was indicative to a CN adduct. The other three diagnostic fragment ions at m/z 274, 246, and 194 are identical to those of nefazodone (Scheme 1).
CM8-CM11 were eluted between 9.9 and 11.9 min and exhibited the identical [M+H]+ at m/z 511, which indicated that they were CN adducts of monohydroxylated nefazodone metabolites. The characteristic fragment ions of m/z 210, 274, and 246 of CM8 revealed that the monohydroxylation took place on the 3-chlorophenylpiperazine ring, which is consistent with previous identified adducts [27, 36]. In comparison, CM9, CM10, and CM11 showed the diagnostic fragment ions at m/z 194, 290, and 262, which are consistent with monohydroxylation on the other side of the molecule. The high abundant CM10 was speculated to contain monohydroxylation on the 5-ethyl side chain, which was in agreement with previously published data [27, 36]. The signal intensity of CM9 and CM11 was much lower than that of CM10, and the current methodology provided sufficient sensitivity for detection of these minor isomers. They were speculated to be minor positional isomers resulting from addition of CN to different positions on the piperazine ring, or potentially from monohydroxylation on positions on the phenoxyethyl-5-ethyltriazolone propyl motif rather than the 5-ethyl side chain (Scheme 1).
CM13 and CM14 were found to be a pair of isomers of CN adducts with [M+H]+ at m/z of 509, two mass unit less than the CN adducts of monohydroxylated nefazodone metabolites. CM13 and CM14 had identical fragment ions at m/z 194 and 288 (Figure 5), indicating further oxidation at the monohydroxylated 5-ethyl side chain to form the ketone form (Scheme 1). The two isomers were proposed to be generated from addition of CN to different positions on the piperazine ring.
CM4 and CM6 showed the identical [M+H]+ at m/z 527, corresponding to CN adducts of dihydroxylated nefazodone metabolites. The fragment ions at m/z 290 and 210 derived from CM4 suggested that the hydroxylation took place on the 3-chlorophenylpiperazine ring and 5-ethyl side chain, respectively. The proposed structure is in consistence with previously reported nefazodone metabolites [27, 36]. In comparison, CM6 gave the fragment ions at m/z 306 and 194 (Supplementary Information, Figure S2), indicating that both hydroxylation events took place on the phenoxyethyl-5-ethyltriazolone propyl motif of the molecule, one presumably being on the 5-ethyl side chain, one on another position.
CM1 and CM2 were proposed to be CN adducts of N-dearylated nefazodone metabolites. CM2 showed [M+H]+ at m/z 399, 14 mass unites greater than CN adduct of N-dearylated nefazodone, consistent with a CN adduct of either a ketone product [36] or an N-methylation product following N-dearylation. The product ion of m/z 274 was identical to that of the parent drug. CM1 was 16 mass units greater than CM2, indicating an additional hydroxylation. Generation of product ion at m/z 290 suggested that the oxygen was added on the 5-ethyl side chain. The product ion at m/z 139 of both adducts suggested that the addition of 14 mass unites took place on the piperazine ring. HR-MS (Orbitrap, Thermo Fisher, Waltham, MA, USA) was applied to determine the elemental composition of these two metabolites. Results suggest that the addition of 14 mass units to CM1 were from the N-methyl group rather than the formation of a ketone (data will be published separately) (Scheme 1). This example showed a significant advantage of HR-MS over Qtrap in metabolite identification.
CM3, CM5, and CM7 were proposed to be a group of CN derivatives of O-dearylated nefazodone. CM3 and CM7 demonstrated [M+H]+ at m/z 435 and 433, respectively. The product ion of m/z 214 of CM3 is consistent with a proposed structure which contains a monohydroxyl group on the 5-ethyl chain, while m/z of 212 of CM7 indicated further oxidation of the monoxdroxyl group to form the ketone structure. CM5 had a [M+H]+ at m/z 447, 14 mass units greater than CM7. Fragment ion at m/z 226 suggested formation of an additional ketone structure. Most likely, the hydroxyl group at CM7 was converted to a carboxylic group in CM5 (Scheme 1).
The current methodology that combined NL scan/filtering of 27 Da, characteristic ER spectrum, and structure-informative EPI spectrum allowed for comprehensive and unambiguous identification of the CN adducts. To our knowledge, eight (CM1, CM2, CM3, CM5, CM6, CM7, CM13, and CM14, Scheme 1) of 14 detected CN adducts were not reported in the literature. In addition, four isomers of CN adducts with a molecular ion at m/z 511 (CM8, CM9, CM10, or CM11) were identified in this study while only two were shown in previous literature. It is worth mentioning that the ER spectrum was very useful for confirmation of the identity of CN adducts. As demonstrated in the examples of CM14 and CM6, their ER spectra showed the typical isotopic pattern when CN and 13C15N were added in equal amount in the microsomal incubation (Figure 5 and Supplementary Information Figure S2). The peak height of the molecular ion of CN derivative to that of the 13C15N is expected to be about 1:1.3, due to the naturally occurring 37Cl isotope. Any detected compound that shows an ER spectrum deviating from the expected pattern is not considered to be a CN adduct. In addition, the EPI scan on Qtrap instrument generated high quality and high abundant spectrum without low mass cut-off. In this case study, the fragments at low molecular weight range were highly useful in providing unequivocal information for identification of the nefazodone metabolites.
3.4 Potential Applications of the Current Workflow
Results from this study demonstrated that current methodology is sensitive and effective in detecting and characterizing GSH and CN adducts, and suitable for high throughput screening of in vitro reactive metabolites in early drug discovery. As shown in Figure 1, the workflow used a generic data acquisition method that combined scans of PI, NL, ER, and EPI with polarity switch so that it can be employed for screening compounds with a variety of structures without prior knowledge of their molecular weights or fragmentation patterns. Recorded EPI data sets that contain product ion spectra of GSH and CN adducts with some interference ions can be processed for selective extraction of GSH and CN adducts and recovery of their product spectra in parallel. The use of stable isotope-labeled GSH and KCN and recording of high quality full scan MS spectra (ER) allowed immediate confirmation of adduct ions and false positives. The generic data acquisition method and fast elimination of false positive are the two key elements of high throughput screening of reactive metabolites. Furthermore, EPI spectrum provided rich fragments without mass cut-off, which is useful in structural elucidation of reactive metabolites. Therefore, LC/MS data sets of a large number of incubation samples can be acquired continuously with the same method. Meantime, post-acquisition data mining can be carried out in parallel. There is no need for a second LC/MS injection for either reactive metabolite detection or recording of their MS/MS spectra.
The work flow displayed in Figure 1 has a great flexibility, enabling screening for either GSH adducts or CN adducts separately, when GSH/stable isotope labeled GSH or KCN/stable isotope labeled KCN is included in the trapping experiment. In addition, this LC/MS method would be suitable for simultaneous screening of GSH and CN adducts without use of stable isotope labeled GSH and KCN, although it may take a longer time to confirm the identity of detected adducts or false positive peaks displayed in NL processed EPI data sets. The same working principle behind the workflow should be applicable to the identification and semiquantification of reactive metabolites trapped by radiolabled GSH, KCN, N-acetyl cysteine and/or cysteine using LC coupled with radiochromatographic detection and Qtrap. It was demonstrated that a method using negative PI to trigger positive EPI was able to sensitively detect N-acetyl cysteine adducts and record their MS/MS spectra [12]. Like GSH adducts, cysteine conjugates underwent a few unique fragmentation pathways in the positive ion mode [23] that can be employed for their detection and MS/MS acquisition by Qtrap instruments.
Like triple quadrupole mass spectrometry, Qtrap mass spectrometry has MRM function that has been used predominantly for quantitation of drugs and metabolites in both in vivo pharmacokinetic studies and in vitro drug metabolism screenings, such as metabolic stability and CYP inhibition assays. On the other hand, ion traps, linear ions traps, and HR-MS instruments are mainly applied for drug metabolite profiling and identifications [39, 40]. However, there is an increased need for the use of a single of LC/MS platform for both bioanalysis and metabolite identification, especially in small discovery organizations. Qtrap has scanning functions of MRM-, NL-, and PI-EPI as well as polarity switching that allow it to be effectively used for profiling and identification of oxidative and conjugated metabolites [41–43], as well as reactive metabolite screening as demonstrated in the literatures [12, 13, 26] and current study. Therefore, Qtrap is well suited for a small discovery or academic drug metabolism and pharmacokinetics research lab where bioanalysis or in vitro quantitative metabolism screening is the major responsibility, while metabolite screening and identification are conducted occasionally. In general, HR-MS has better capabilities in drug metabolite profiling and identification than Qtrap. However, it is not practical to equip two or multiple LC/MS instruments in a small lab, each of which is dedicated to either quantitative or qualitative analysis. HR-MS is a preferred LC/MS platform for a drug metabolism lab where metabolite identification is its major task, while quantitative analysis in discovery pharmacokinetic and in vitro metabolism studies is performed occasionally [44, 45].
4 Conclusion
It is important to screen both hard and soft reactive metabolites for a comprehensive evaluation of lead compounds for their bioactivation potentials. Traditional methods usually involve separate trapping experiments and discrete LC-MS/MS analyses in order to cover all possible bioactivation pathways. In the current study, we took advantage of the unique scanning function and capability of the API Qtrap 5500 to establish a method for simultaneous detection and identification of both soft and hard reactive metabolites. The trapping agents for soft and hard reactive metabolites, GSH and KCN, as well as their stable isotope labeled versions, were included in the same incubations. Negative PI scan of m/z 272 for detection of GSH adducts and positive NL scans of 27 and 29 for detection of CN adducts were conducted as survey scans to trigger acquisition of ER and EPI spectra in positive mode. Due to the superior acquisition speed and fast polarity switch, multiple types of scans, including PI scan of m/z 272, NL scan of 27 Da, NL of 29 Da, ER, and EPI, as well as a polarity switch were completed in a single run. Post-acquisition data mining of EPI spectrum using NL filter of 129 and 27 Da were then utilized to more selectively reveal GSH adducts and CN adducts, respectively. EPI spectrum provided high quality fragmentation ions without one-third low mass cut-off, which are very informative for structure elucidation. In addition, the isotope pattern in the ER spectrum was also inspected for each of the identified peaks to exclude any potential false positives. By using current approach, six GSH adduct and 14 CN adducts of nefazodone were unambiguously detected and structurally characterized in a single run, some of which were not reported in the literature. The results suggested that utilization of multiple types of scans (PI, NL, ER, and EPI) enabled highly selective and fast identification of the reactive metabolites, with superior sensitivity for the minor ones. Overall, the current approach is very efficient in the detection and structural characterization of both soft and hard reactive metabolites in a single LC-MS/MS run. It can be used as a high-throughput method to comprehensively screen compounds for their bioactivation potentials in early drug discovery. Like triple quadrupole MS instrument, Qtrap has been widely employed for quantitation of drugs and metabolites in both in vivo pharmacokinetic study and in vitro drug metabolism screening, such as metabolic stability and CYP inhibition assays. The drug metabolite identification capability of Qtrap that is demonstrated in the literature [34] and in the current study allows Qtrap to be used as a single LC/MS platform for quantitative and qualitative analysis in discovery drug metabolism and pharmacokinetics studies.
References
Pearson, R.G.: Hard and soft acids and bases. J. Am. Chem. Chem. 85, 3533–3539 (1963)
Kalgutkar, A.S.: In: Lee, M.S., Zhu, M. (eds.) Metabolic Activation of Organic Functional Groups Utilized in Medicinal Chemistry. Mass Spectrometry in Drug Metabolism and Disposition: Basic Principles and Applications, p. 43. John Wiley & Sons, Inc, Hoboken, NJ (2011)
Zhang, Z., Chen, Q., Li, Y., Doss, G.A., Dean, B.J., Ngui, J.S., Silva Elipe, M., Kim, S., Wu, J.Y., Dininno, F., Hammond, M.L., Stearns, R.A., Evans, D.C., Baillie, T.A., Tang, W.: In vitro bioactivation of dihydrobenzoxathiin selective estrogen receptor modulators by cytochrome P450 3A4 in human liver microsomes: formation of reactive iminium and quinone type metabolites. Chem. Res. Toxicol. 18, 675–685 (2005)
Prakash, C., Sharma, R., Gleave, M., Nedderman, A.: In vitro screening techniques for reactive metabolites for minimizing bioactivation potential in drug discovery. Curr. Drug Metab. 9, 952–964 (2008)
Wen, B., Fitch, W.L.: Analytical strategies for the screening and evaluation of chemically reactive drug metabolites. Expert Opin Drug Metab. Toxicol. 5, 39–55 (2009)
Ma, S., Zhu, M.: Recent advances in applications of liquid chromatography-tandem mass spectrometry to the analysis of reactive drug metabolites. Chem. Biol. Interact. 179, 25–37 (2009)
Stepan, A.F., Walker, D.P., Bauman, J., Price, D.A., Baillie, T.A., Kalgutkar, A.S., Aleo, M.D.: Structural Alert/Reactive Metabolite Concept as Applied in Medicinal Chemistry to Mitigate the Risk of Idiosyncratic Drug Toxicity: A Perspective Based on the Critical Examination of Trends in the Top 200 Drugs Marketed in the United States. Chem. Res. Toxicol. (2011), Epub ahead of print.
Humphreys, W.G.: Overview of strategies for addressing BRIs in drug discovery: Impact on optimization and design. Chem. Biol. Interact. 192, 56–59 (2011)
Thompson, R.A., Isin, E.M., Li, Y., Weaver, R., Weidolf, L., Wilson, I., Claesson, A., Page, K., Dolgos, H., Kenna, J.G.: Risk assessment and mitigation strategies for reactive metabolites in drug discovery and development. Chem. Biol. Interact. 192, 65–71 (2010)
Zhuo, X., Hartz, R.A., Bronson, J.J., Wong, H., Ahuja, V.T., Vrudhula, V.M., Leet, J.E., Huang, S., Macor, J.E., Shu, Y.Z.: Comparative biotransformation of pyrazinone-containing corticotropin-releasing factor receptor-1 antagonists: Minimizing the reactive metabolite formation. Drug Metab. Dispos. 38, 5–15 (2010)
Lim, H.K., Chen, J., Sensenhauser, C., Cook, K., Preston, R., Thomas, T., Shook, B., Jackson, P.F., Rassnick, S., Rhodes, K., Gopaul, V., Salter, R., Silva, J., Evans, D.C.: Overcoming the genotoxicity of a pyrrolidine substituted arylindenopyrimidine as a potent dual adenosine A(2A)/A(1) antagonist by minimizing bioactivation to an iminium ion reactive intermediate. Chem. Res. Toxicol. 24, 1012–1030 (2011)
Jian, W., Yao, M., Zhang, D., Zhu, M.: Rapid detection and characterization of in vitro and urinary N-acetyl-L-cysteine conjugates using quadrupole-linear ion trap mass spectrometry and polarity switching. Chem. Res. Toxicol. 22, 1246–1255 (2009)
Wen, B., Ma, L., Nelson, S.D., Zhu, M.: High-throughput screening and characterization of reactive metabolites using polarity switching of hybrid triple quadrupole linear ion trap mass spectrometry. Anal. Chem. 80, 1788–1799 (2008)
Gan, J., Harper, T.W., Hsueh, M.M., Qu, Q., Humphreys, W.G.: Dansyl glutathione as a trapping agent for the quantitative estimation and identification of reactive metabolites. Chem. Res. Toxicol. 18, 896–903 (2005)
Leblanc, A., Shiao, T.C., Roy, R., Sleno, L.: Improved detection of reactive metabolites with a bromine-containing glutathione analog using mass defect and isotope pattern matching. Rapid Commun. Mass Spectrom. 24, 1241–1250 (2010)
Soglia, J.R., Contillo, L.G., Kalgutkar, A.S., Zhao, S., Hop, C.E., Boyd, J.G., Cole, M.J.: A semiquantitative method for the determination of reactive metabolite conjugate levels in vitro utilizing liquid chromatography-tandem mass spectrometry and novel quaternary ammonium glutathione analogues. Chem. Res. Toxicol. 19, 480–490 (2006)
Ma, L., Wen, B., Ruan, Q., Zhu, M.: Rapid screening of glutathione-trapped reactive metabolites by linear ion trap mass spectrometry with isotope pattern-dependent scanning and post-acquisition data mining. Chem. Res. Toxicol. 21, 1477–1483 (2008)
Yan, Z., Caldwell, G.W.: Stable-isotope trapping and high-throughput screenings of reactive metabolites using the isotope MS signature. Anal. Chem. 76, 6835–6847 (2004)
Yan, Z., Caldwell, G.W., Maher, N.: Unbiased high-throughput screening of reactive metabolites on the linear ion trap mass spectrometer using polarity switch and mass tag triggered data-dependent acquisition. Anal. Chem. 80, 6410–6422 (2008)
Lim, H.K., Chen, J., Cook, K., Sensenhauser, C., Silva, J., Evans, D.C.: A generic method to detect electrophilic intermediates using isotopic pattern triggered data-dependent high-resolution accurate mass spectrometry. Rapid Commun. Mass Spectrom. 22, 1295–1311 (2008)
Evans, D.C., Watt, A.P., Nicoll-Griffith, D.A., Baillie, T.A.: Drug-protein adducts: an industry perspective on minimizing the potential for drug bioactivation in drug discovery and development. Chem. Res. Toxicol. 17, 3–16 (2004)
Meneses-Lorente, G., Sakatis, M.Z., Schulz-Utermoehl, T., De Nardi, C., Watt, A.P.: A quantitative high-throughput trapping assay as a measurement of potential for bioactivation. Anal. Biochem. 351, 266–272 (2006)
Inoue, K., Shibata, Y., Takahashi, H., Ohe, T., Chiba, M., Ishii, Y.: A trapping method for semi-quantitative assessment of reactive metabolite formation using [35S]cysteine and [14C]cyanide. Drug Metab. Pharmacokinet. 24, 245–254 (2009)
Levsen, K., Schiebel, H.M., Behnke, B., Dotzer, R., Dreher, W., Elend, M., Thiele, H.: Structure elucidation of phase II metabolites by tandem mass spectrometry: An overview. J Chromatogr. A 1067, 55–72 (2005)
Dieckhaus, C.M., Fernandez-Metzler, C.L., King, R., Krolikowski, P.H., Baillie, T.A.: Negative ion tandem mass spectrometry for the detection of glutathione conjugates. Chem. Res. Toxicol. 18, 630–638 (2005)
Zheng, J., Ma, L., Xin, B., Olah, T., Humphreys, W.G., Zhu, M.: Screening and identification of GSH-trapped reactive metabolites using hybrid triple quadruple linear ion trap mass spectrometry. Chem. Res. Toxicol. 20, 757–766 (2007)
Argoti, D., Liang, L., Conteh, A., Chen, L., Bershas, D., Yu, C.P., Vouros, P., Yang, E.: Cyanide trapping of iminium ion reactive intermediates followed by detection and structure identification using liquid chromatography-tandem mass spectrometry (LC-MS/MS). Chem. Res. Toxicol. 18, 1537–1544 (2005)
Zhu, M., Ma, L., Zhang, H., Humphreys, W.G.: Detection and structural characterization of glutathione-trapped reactive metabolites using liquid chromatography-high-resolution mass spectrometry and mass defect filtering. Anal. Chem. 79, 8333–8341 (2007)
Barbara, J.E., Castro-Perez, J.M.: High-resolution chromatography/time-of-flight MSE with in silico data mining is an information-rich approach to reactive metabolite screening. Rapid Commun. Mass Spectrom. 25, 3029–3040 (2011)
Rousu, T., Pelkonen, O., Tolonen, A.: Rapid detection and characterization of reactive drug metabolites in vitro using several isotope-labeled trapping agents and ultra-performance liquid chromatography/time-of-flight mass spectrometry. Rapid Commun. Mass Spectrom. 23, 843–855 (2009)
Mitchell, M.D., Elrick, M.M., Walgren, J.L., Mueller, R.A., Morris, D.L., Thompson, D.C.: Peptide-based in vitro assay for the detection of reactive metabolites. Chem. Res. Toxicol. 21, 859–868 (2008)
Yan, Z., Maher, N., Torres, R., Huebert, N.: Use of a trapping agent for simultaneous capturing and high-throughput screening of both "soft" and "hard" reactive metabolites. Anal. Chem. 79, 4206–4214 (2007)
Baillie, T.A., Davis, M.R.: Mass spectrometry in the analysis of glutathione conjugates. Biol. Mass Spectrom. 22, 319–325 (1993)
Jian, W., Yao, M., Wen, B., Jones, E.B., Zhu, M.: In: Lee, M.S., Zhu, M. (eds.) Use of triple quadrupole-linear ion trap mass spectrometry as a single LC/MS platform in drug metabolism and pharmacokinetics studies. Mass Spectrometry in Drug Metabolism and Disposition: Basic Principles and Applications, pp. 483–524. John Wiley and Sons, Inc, Hoboken, NJ (2011)
Kalgutkar, A.S., Vaz, A.D., Lame, M.E., Henne, K.R., Soglia, J., Zhao, S.X., Abramov, Y.A., Lombardo, F., Collin, C., Hendsch, Z.S., Hop, C.E.: Bioactivation of the nontricyclic antidepressant nefazodone to a reactive quinone-imine species in human liver microsomes and recombinant cytochrome P450 3A4. Drug Metab. Dispos. 33, 243–253 (2005)
Bauman, J.N., Frederick, K.S., Sawant, A., Walsky, R.L., Cox, L.M., Obach, R.S., Kalgutkar, A.S.: Comparison of the bioactivation potential of the antidepressant and hepatotoxin nefazodone with aripiprazole, a structural analog and marketed drug. Drug Metab. Dispos. 36, 1016–1029 (2008)
Li, A.C., Shou, W.Z., Mai, T.T., Jiang, X.Y.: Complete profiling and characterization of in vitro nefazodone metabolites using two different tandem mass spectrometric platforms. Rapid Commun. Mass Spectrom. 21, 4001–4008 (2007)
Peterman, S.M., Duczak, N., Kalgutkar, A.S., Lame, M.E., Soglia, J.R.: Application of a linear ion trap/Orbitrap mass spectrometer in metabolite characterization studies: Examination of the human lever microsomal metabolism of the non-tricyclic anti-depressant nefazodone using data-dependent accurate mass measurements. J. Am. Soc. Mass Spectrom. 17, 363–375 (2006)
Prakash, C., Shaffer, C.L., Nedderman, A.: Analytical strategies for identifying drug metabolites. Mass Spectrom. Rev. 26, 340–369 (2007)
Zhu, M., Zhang, H., Humphreys, W.G.: Drug metabolite profiling and identification by high-resolution mass spectrometry. J. Biol. Chem. 286, 25419–25425 (2011)
Mauriala, T., Chauret, N., Oballa, R., Nicoll-Griffith, D.A., Bateman, K.P.: A strategy for identification of drug metabolites from dried blood spots using triple-quadrupole/linear ion trap hybrid mass spectrometry. Rapid Commun. Mass Spectrom. 19, 1984–1992 (2005)
Yao, M., Ma, L., Humphreys, W.G., Zhu, M.: Rapid screening and characterization of drug metabolites using a multiple ion monitoring-dependent MS/MS acquisition method on a hybrid triple quadrupole-linear ion trap mass spectrometer. J. Mass Spectrom. 43, 1364–1375 (2008)
Xia, Y.Q., Miller, J.D., Bakhtiar, R., Franklin, R.B., Liu, D.Q.: Use of a quadrupole linear ion trap mass spectrometer in metabolite identification and bioanalysis. Rapid Commun. Mass Spectrom. 17, 1137–1145 (2003)
Bateman, K.P., Kellmann, M., Muenster, H., Papp, R., Taylor, L.: Quantitative-qualitative data acquisition using a benchtop Orbitrap mass spectrometer. J. Am. Soc. Mass Spectrom. 20, 1441–1450 (2009)
Ramanathan, R., Josephs, J.L., Jemal, M., Arnold, M., Humphreys, W.G.: Novel MS solutions inspired by MIST. Bioanalysis 2, 1291–1313 (2010)
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
Figure S1
Positive ER spectrum (A), positive EPI spectra (B), and proposed structure of nefazodone GSH adduct GM5. (DOC 114 kb)
Figure S2
Positive ER spectrum (A), positive EPI spectra (B), and proposed structure of nefazodone CN adduct CM6. (DOC 87 kb)
Rights and permissions
About this article
Cite this article
Jian, W., Liu, HF., Zhao, W. et al. Simultaneous Screening of Glutathione and Cyanide Adducts Using Precursor Ion and Neutral Loss Scans-Dependent Product Ion Spectral Acquisition and Data Mining Tools. J. Am. Soc. Mass Spectrom. 23, 964–976 (2012). https://doi.org/10.1007/s13361-012-0354-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-012-0354-6