Abstract
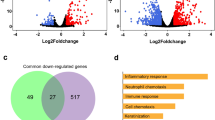
tsRNAs (tRNA-derived small non-coding RNAs), including tRNA halves (tiRNAs) and tRNA fragments (tRFs), have been implicated in some viral infections, such as respiratory viral infections. However, their involvement in SARS-CoV infection is completely unknown. A comprehensive analysis was performed to determine tsRNA populations in a mouse model of SARS-CoV-infected samples containing the wild-type and attenuated viruses. Data from the Gene Expression Omnibus (GEO) dataset at NCBI (accession ID GSE90624) was used for this study. A count matrix was generated for the tRNAs. Differentially expressed tRNAs, followed by tsRNAs derived from each significant tRNAs at different conditions and time points between the two groups WT(SARS-CoV-MA15-WT) vs Mock and ΔE (SARS-CoV-MA15-ΔE) vs Mock were identified. Notably, significantly differentially expressed tRNAs at 2dpi but not at 4dpi. The tsRNAs originating from differentially expressed tRNAs across all the samples belonging to each condition (WT, ΔE, and Mock) were identified. Intriguingly, tRFs (tRNA-derived RNA fragments) exhibited higher levels compared to tiRNAs (tRNA-derived stress-induced RNAs) across all samples associated with WT SARS-CoV strain compared to ΔE and mock-infected samples. This discrepancy suggests a non-random formation of tsRNAs, hinting at a possible involvement of tsRNAs in SARS-CoV viral infection.







Similar content being viewed by others
Data availability
A publicly available dataset was analyzed in this study. This data can be found in the link https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE90624
References
Alexandrov A, Chernyakov I, Gu W et al (2006) Rapid tRNA decay can result from lack of nonessential modifications. Mol Cell 21:87–96. https://doi.org/10.1016/J.MOLCEL.2005.10.036
Anderson P, Ivanov P (2014) tRNA fragments in human health and disease. FEBS Lett 588:4297. https://doi.org/10.1016/J.FEBSLET.2014.09.001
Arimbasseri AG, Maraia RJ (2016) RNA polymerase III Advances: structural and tRNA functional views. Trends Biochem Sci 41:546. https://doi.org/10.1016/J.TIBS.2016.03.003
Bartel DP (2004) MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116:281–297. https://doi.org/10.1016/S0092-8674(04)00045-5
Bidet K, Dadlani D, Garcia-Blanco MA (2014) G3BP1, G3BP2 and CAPRIN1 are required for translation of interferon stimulated mRNAs and are targeted by a dengue virus non-coding RNA. PLoS Pathog 10:e1004242. https://doi.org/10.1371/JOURNAL.PPAT.1004242
Chan PP, Lowe TM (2016) GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44:D184–D189. https://doi.org/10.1093/NAR/GKV1309
Damas ND, Fossat N, Scheel TKH (2019) Functional interplay between RNA viruses and non-coding RNA in mammals. Noncoding RNA 5:7. https://doi.org/10.3390/NCRNA5010007
DeDiego ML, Álvarez E, Almazán F et al (2007) A severe acute respiratory syndrome coronavirus that lacks the E gene is attenuated in vitro and in vivo. J Virol 81:1701. https://doi.org/10.1128/JVI.01467-06
Deng J, Ptashkin RN, Chen Y et al (2015) Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Molecular Therapy 23:1622. https://doi.org/10.1038/MT.2015.124
Emara MM, Ivanov P, Hickman T et al (2010) Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J Biol Chem 285:10959–10968. https://doi.org/10.1074/jbc.M109.077560
Fu H, Feng J, Liu Q et al (2009) Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett 583:437–442. https://doi.org/10.1016/J.FEBSLET.2008.12.043
Gebert D, Hewel C, Rosenkranz D (2017) unitas: the universal tool for annotation of small RNAs. BMC Genomics 18:1–14. https://doi.org/10.1186/S12864-017-4031-9
Grundhoff A, Sullivan CS (2011) Virus-encoded microRNAs. Virology 411:325. https://doi.org/10.1016/J.VIROL.2011.01.002
Haiser HJ, Karginov FV, Hannon GJ, Elliot MA (2008) Developmentally regulated cleavage of tRNAs in the bacterium Streptomyces coelicolor. Nucleic Acids Res 36:732–741. https://doi.org/10.1093/NAR/GKM1096
Hunter JD (2007) Matplotlib: a 2D graphics environment. Comput Sci Eng 9:90–95. https://doi.org/10.1109/MCSE.2007.55
Ibba M, Söll D (2004) Aminoacyl-tRNAs: setting the limits of the genetic code. Genes Dev 18:731–738. https://doi.org/10.1101/GAD.1187404
Jiang H, Wong WH (2008) SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics 24:2395. https://doi.org/10.1093/BIOINFORMATICS/BTN429
Kumar P, Anaya J, Mudunuri SB, Dutta A (2014) Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biol 12:1–14. https://doi.org/10.1186/S12915-014-0078-0
Larminie CGC, Sutcliffe JE, Tosh K et al (1999) Activation of RNA polymerase III transcription in cells transformed by simian virus 40. Mol Cell Biol 19:4927. https://doi.org/10.1128/MCB.19.7.4927
Lee SR, Collins K (2005) Starvation-induced cleavage of the tRNA anticodon loop in Tetrahymena thermophila. J Biol Chem 280:42744–42749. https://doi.org/10.1074/JBC.M510356200
Lee YS, Shibata Y, Malhotra A, Dutta A (2009) A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev 23:2639. https://doi.org/10.1101/GAD.1837609
Li S, Xu Z, Sheng J (2018) tRNA-derived small RNA: a novel regulatory small non-coding RNA. Genes (Basel) 9:246. https://doi.org/10.3390/GENES9050246
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:1–21. https://doi.org/10.1186/S13059-014-0550-8
Morales L, Oliveros JC, Fernandez-Delgado R et al (2017) SARS-CoV-encoded small RNAs contribute to infection-associated lung pathology. Cell Host Microbe 21:344. https://doi.org/10.1016/J.CHOM.2017.01.015
Pan T (2018) Modifications and functional genomics of human transfer RNA. Cell Res 28:395–404. https://doi.org/10.1038/S41422-018-0013-Y
Perez JT, Varble A, Sachidanandam R et al (2010) Influenza A virus-generated small RNAs regulate the switch from transcription to replication. Proc Natl Acad Sci U S A 107:11525–11530. https://doi.org/10.1073/PNAS.1001984107/-/DCSUPPLEMENTAL
Roberts A, Deming D, Paddock CD et al (2007) A mouse-adapted SARS-coronavirus causes disease and mortality in BALB/c mice. PLoS Pathog 3:0023–0037. https://doi.org/10.1371/JOURNAL.PPAT.0030005
Ruggero K, Guffanti A, Corradin A et al (2014) Small noncoding RNAs in cells transformed by human T-cell leukemia virus type 1: a role for a tRNA fragment as a primer for reverse transcriptase. J Virol 88:3612. https://doi.org/10.1128/JVI.02823-13
Ruivinho C, Gama-Carvalho M (2023) Small non-coding RNAs encoded by RNA viruses: old controversies and new lessons from the COVID-19 pandemic. Front Genet 14:1216890. https://doi.org/10.3389/FGENE.2023.1216890/BIBTEX
Selitsky SR, Baran-Gale J, Honda M et al (2015) Small tRNA-derived RNAs are increased and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep 5:1–7. https://doi.org/10.1038/srep07675
Shi J, Zhang Y, Zhou T, Chen Q (2019) tsRNAs: the Swiss Army Knife for translational regulation. Trends Biochem Sci 44:185–189. https://doi.org/10.1016/J.TIBS.2018.09.007
Shigematsu M, Kirino Y (2015) tRNA-derived short non-coding RNA as interacting partners of Argonaute proteins. Gene Regul Syst Bio 9:27. https://doi.org/10.4137/GRSB.S29411
Thompson DM, Parker R (2009) Stressing out over tRNA cleavage. Cell 138:215–219. https://doi.org/10.1016/J.CELL.2009.07.001
Tycowski KT, Guo YE, Lee N et al (2015) Viral noncoding RNAs: more surprises. Genes Dev 29:567. https://doi.org/10.1101/GAD.259077.115
Vaňáčová Š, Wolf J, Martin G et al (2005) A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol 3:e189. https://doi.org/10.1371/JOURNAL.PBIO.0030189
Wang Q, Lee I, Ren J et al (2013) Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Molecular Therapy 21:368. https://doi.org/10.1038/MT.2012.237
Weng KF, Hung CT, Hsieh PT et al (2014) A cytoplasmic RNA virus generates functional viral small RNAs and regulates viral IRES activity in mammalian cells. Nucleic Acids Res 42:12789. https://doi.org/10.1093/NAR/GKU952
Yamasaki S, Ivanov P, Hu GF, Anderson P (2009) Angiogenin cleaves tRNA and promotes stress-induced translational repression. J Cell Biol 185:35. https://doi.org/10.1083/JCB.200811106
Yu X, Xie Y, Zhang S et al (2021) tRNA-derived fragments: mechanisms underlying their regulation of gene expression and potential applications as therapeutic targets in cancers and virus infections. Theranostics 11:461–469. https://doi.org/10.7150/thno.51963
Acknowledgements
The authors acknowledge the support extended by the Indian Council of Medical Research (ICMR), New Delhi, and All India Institute of Medical Sciences (AIIMS), New Delhi, for providing computational facilities.
Funding
Research reported in this study was supported by the Indian Council of Medical Research (ICMR) under award number BMI/11(36)/2022 to Swati Ajmeriya.
Author information
Authors and Affiliations
Contributions
S.A. designed the analysis, applied methods, and wrote the main manuscript; D.B. applied methods, and reviewed the manuscript; A.K. applied methods and edited and reviewed the manuscript; H.S. reviewed and edited the manuscript; S.R. edited the manuscript. S.K. supervised and reviewed the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethical approval
This declaration is not applicable
Competing interests
The authors declare no competing interests.
Additional information
Communicated by: Agnieszka Szalewska-Palasz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ajmeriya, S., Bharti, D.R., Kumar, A. et al. In silico approach for the identification of tRNA-derived small non-coding RNAs in SARS-CoV infection. J Appl Genetics 65, 403–413 (2024). https://doi.org/10.1007/s13353-024-00853-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-024-00853-4