Abstract
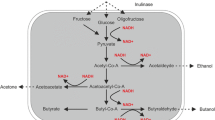
The rapid development of new molecular methods and approaches, sequencing technologies, has provided new insights into genetic and structural features of bacterial genomes. Information about the genetic organization of metabolic pathways and their regulatory elements has greatly contributed to the increase in the number of studies related to the construction of new bacterial strains with improved characteristics. In this study, the entire genome of the producing strain Clostridium sp. UCM В-7570 from the “Collection of producing strains of microorganisms and plant lines for food and agricultural biotechnology” of Institute of Food Biotechnology and Genomics of the National Academy of Sciences of Ukraine was sequenced and characterized. The genome was assembled into the scaffold with a total size of 4,470,321 bp and a GC content of 29.7%. The total number of genes identified was 4262, of which 4057 encoded proteins, 10 were rRNA operons, and 80 were tRNA genes. The genes of the sequenced genome encoding enzymes involved in butanol fermentation were found and analyzed. They were organized into cluster structures, and their protein sequences were found to be similar to the corresponding strains of C. acetobutylicum, C. beijerinckii, and C. pasteurianum type strains with the highest similarity to the latter. Thus, Clostridium sp. UCM В-7570 producing strain was identified as C. pasteurianum and suggested for metabolic engineering purposes.





Similar content being viewed by others
References
Abdelaal AS, Yazdani SS (2022) Engineering E.coli to synthesize butanol. Biochem Soc 50(20):867–876. https://doi.org/10.1042/BST20211009
Arbter P, Sabra W, Utesch T, Hong Y, Zeng A-P (2021) Metabolomic and kinetic investigations on the electricity-aided production of butanol by Clostridium pasteurianum strains. Eng Life Sci 21:181–195. https://doi.org/10.1002/elsc.202000035
Biebl H (2001) Fermentation of glycerol by Clostridium pasteurianum - batch and continuous culture studies. J Ind Microbiol Biotechnol 27:18–26. https://doi.org/10.1038/sj.jim.7000155
Chen K-T, Liu C-L, Huang S-H, Shen H-T, Shieh Y-K, Chiu H-A, Lu CL (2018a) CSAR: a contig scaffolding tool using algebraic rearrangements. Bioinformatics 34(1):109–111. https://doi.org/10.1093/bioinformatics/btx543
Chen K-T, Shen H-T, Lu CL (2018b) Multi-CSAR: a multiple reference-based contig scaffolder using algebraic rearrangements. BMC Syst Biol 12(S9):139. https://doi.org/10.1186/s12918-018-0654-y
Edwards K, Logan J, Saunders N (2005) Real-time PCR: an essential guide. Emerg Infect Dis 11(1):185–186. https://doi.org/10.3201/eid1101.040896
Gallardo R, Alves M, Rodrigues LR (2014) Modulation of crude glycerol fermentation by Clostridium pasteurianum DSM 525 towards the production of butanol. Biomass and Bioenergy 71:134–143. https://doi.org/10.1016/j.biombioe.2014.10.015
Hussain A, Shahbaz U, Khan S, Basharat S, Ahmad K, Khan F, Xia X (2022) Advances in microbial metabolic engineering for the production of butanol isomers (isobutanol and 1-butanol) from a various biomass. Bioenerg Res. https://doi.org/10.1007/s12155-022-10410-8
Hyatt D, Chen G-L, LoCascio PF, Land ML, Larimer FW, Hauser LJ (2010) Prodigal: prokaryotic gene recognition and translation initiation site identification. BMC Bioinf 11(119). https://doi.org/10.1186/1471-2105-11-119
Janda JM, Abbott SL (2007) 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol 45(9):2761–2764. https://doi.org/10.1128/JCM.01228-07
Jiang Y, Wu R, Lu J, Dong W, Zhou J, Zhang W, Xin F, Jiang M (2021) Quantitative proteomic analysis to reveal expression difference for butanol production from glycerol and glucose by Clostridium sp. stain CT7. Microb Cell Fact 20(12). https://doi.org/10.1186/s12934-021-01508-3
Keis S, Bennett CF, Ward KV, Jones DT (1995) Taxonomy and phylogeny of industrial solvent-producing clostridia. J of System Bacteriol 45(4):693–705. https://doi.org/10.1099/00207713-45-4-693
Kumar CM, Mugunthan M, Kapoor CR, Pandalanghat CS (2019) Speciation of fungi using real time PCR with molecular beacons: can we solve the enigma of diagnosis of invasive fungal disease. Med J Armed Forces India 75:41–49. https://doi.org/10.1016/j.mjafi.2017.12.003
Landlinger C, Preuner S, Baskova L, van Grotel M, Hartwig NG, Dworzak M, Mann G, Attarbaschi A, Kager L, Peters C, Matthes-Martin S, Lawitschka A, van den Heuvel-Eibrink MM, Lion T (2010) Diagnosis of invasive fungal infections by a realtime panfungal PCR assay in immunocompromised pediatric patients. Leukemia 24(12):2032–2038. https://doi.org/10.1038/leu.2010.20933
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23:2947–2948. https://doi.org/10.1093/bioinformatics/btm404
Li H, Durbin R, Notes A (2010) Fast and accurate long-read alignment with Burrowa-Wheeler transform. Bioiformatics 26(5):589–595. https://doi.org/10.1093/bioinformatics/btp698
Li W, O'Neill KR, Haft DH, DiCuccio M, Chetvernin V, Badretdin A, Coulouris G, Chitsaz F, Derbyshire MK, Durkin AS, Gonzales NR, Gwadz M, Lanczycki CJ, Song JS, Thanki N, Wang J, Yamashita RA, Yang M, Zheng C et al (2021) RefSeq: expanding the Prokaryotic Genome Annotation Pipeline reach with protein family model curation. Nucleic Acids Res 49(D1):D1020–D1028. https://doi.org/10.1093/nar/gkaa1105
Lin S-D, Yen H-W, Kao W-C, Cheng C-L, Chen W-M, Huang C-C, Chang J-S (2015) Bio-butanol production from glycerol with Clostridium pasteurianum CH4: the effects of butyrate addition and in situ butanol removal via membrane distillation. Biotech of biofuels 8:168. https://doi.org/10.1186/s13068-015-0352-6
Liberato V, Benevenuti C, Coelho F, Botelho A, Amaral P, NJr P, Ferreira T (2019) Clostridium sp. as a bio-catalyst for fuels and chemicals production in a biorefinery context. Catalysts 9(962):1–37. https://doi.org/10.3390/catal9110962
Malaviya A, Jang YS, Lee SY (2012) Continuous butanol production with reduced byproducts formation from glycerol by a hyper producing mutant of Clostridium pasteurianum. Appl Microbiol Biotechnol 93:1485–1494. https://doi.org/10.1007/s00253-011-3629-0
Menzel P, Ng K, Krogh A (2016) Fast and sensitive taxonomic classification for metagenomics with Kaiju. Nat Commun 7:11257. https://doi.org/10.1038/ncomms11257
Millon L, Larosa F, Lepiller Q, Legrand F, Rocchi S, Daguindau E, Scherer E, Bellanger A-P, Leroy J, Grenouillet F (2013) Quantitative polymerase chain reaction detection of circulating DNA in serum for early diagnosis of mucormycosis in immunocompromised patients. Clin Infect Dis 56(10):95–101. https://doi.org/10.1093/cid/cit094
Munch G, Mittler J, Rehmann L (2020) Increased selectivity for butanol in Clostridium pasteurianum fermentations via butyric acid addition or dual feedstock strategy. Fermentation 6(3):67. https://doi.org/10.3390/fermentation6030067
Nederbragt AJ (2014) On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol 15(113). https://doi.org/10.1186/gb4173
Ondov BD, Bergman NH, Phillippy AM (2011) Interactive metagenomic visualization in a web browser. BMC Bioinformatics 12:385. https://doi.org/10.1186/1471-2105-12-385
Poehlein A, Grosse-Honebrink A, Zhang Y, Minton NP, Daniel R (2015) Complete genome sequence of the nitrogen-fixing and solvent-producing Clostridium pasteurianum DSM 525. Genome Announc 3:e01591–e01514. https://doi.org/10.1128/genomeA.01591-14
Potschka A, Bock HG (2021) Correction to: a sequential homotopy method for mathematical programming problems. Math Program 190:843–844. https://doi.org/10.1007/s10107-021-01668-5
Pyne ME, Moo-Young M, Chung DA, Chou CP (2013) Development of an electrotransformation protocol for genetic manipulation of Clostridium pasteurianum. Biotechnology for biofuels. 6(1):1–20. https://doi.org/10.1186/1754-6834-6-50
Pyne ME, Liu X, Moo-Young M, Chung DA, Chou P (2016) Genome-directed analysis of prophage excision, host defence systems, and central fermentative metabolism in Clostridium pasteurianum. Sci Rep 6:26228
Radchenko M, Tigunova O, Zelena L, Beiko N, Andriiash H, Shulga S (2021) Phylogenetic analysis of the Bacillus subtilis IFBG MK-2 strain and riboflavin production by its induced clones. Cytol Genet 55(2):145–151. https://doi.org/10.3103/S0095452721020134
Rappert S, Song L, Sabra W, Wang W, Zeng A-P (2013) Draft genome sequence of type strain Clostridium pasteurianum DSM 525 (ATCC 6013), a promising producer of chemicals and fuels. Genome Announc 1:e00232–e00212. https://doi.org/10.1128/genomeA.00232-12
Ren H, Hu H, Zeng Z (2021) Sequential probabilistic ratio test for the scale parameter of the ρ-norm distribution. Discrete Dyn Nat Soc 2021:7. https://doi.org/10.1155/2021/9922435
Rotta C, Poehlein A, Schwarz K, McClure P, Daniel R, Minton NP (2015) Closed genome sequence of Clostridium pasteurianum ATCC 6013. Genome Announc 3:e01596–e01514. https://doi.org/10.1128/genomeA.01596-14
Sandoval NR, Venkataramanan KP, Groth TS, Papoutsakis ET (2015) Whole-genome sequence of an evolved Clostridium pasteurianum strain reveals Spo0A deficiency responsible for increased butanol production and superior growth. Biotechnol Biofuels 8:227. https://doi.org/10.1186/s13068-015-0408-7
Sarchami T, Munch G, Johnson E, Kieblich S, Rehmann L (2016) A review of process-design challenges for industrial fermentation of butanol from crude glycerol by non-biphasic Clostridium pasteurianum. Fermentation 2(2):13. https://doi.org/10.3390/fermentation2020013
Shekelyan M, Cormode G (2021) Sequential random sampling revisited: hidden shuffle method. Proceedings of The 24th International Conference on Artificial Intelligence and Statistics 130:3628-3636 https://proceedings.mlr.press/v130/shekelyan21a.html.
Schwarz KM, Grosse-Honebrink A, Derecka K, Rotta C, Zhang Y, Minton NP (2017) Towards improved butanol production through targeted genetic modification of Clostridium pasteurianum. Metab Eng 40:124–137. https://doi.org/10.1016/j.ymben.2017.01.009
Tigunova OO, Beiko NY, Andriiash HS, Pryiomov SH, Shulga SM (2017) Domestic butanol-producing strains of the Clostridium genus. Biotechnologia Acta 10(1):34–42. https://doi.org/10.15407/biotech10.01.034
Tigunova O, Shulga S, Blume Y (2013) Biobutanol as an alternative type of fuel. Cytol Genet 47(6):366–382. https://doi.org/10.3103/s0095452713060042
Wilkinson SR, Young M, Goodacre R, Morris JG, Farrow JAE, Collins MD (1995) Phenotypic and genotypic differences between certain strains of Clostridium acetobutylicum. FEMS Microbiol Lett 125:199–204. https://doi.org/10.1111/j.1574-6968.1995.tb07358.x
Wilson K (2001) Preparation of genomic DNA from bacteria. Curr protoc. Mol Biol:2.4.1–2.4.5. https://doi.org/10.1002/0471142727.mb0204s56
Xin F, Wang C, Dong W, Zhang W, Wu H, Ma J, Jang M (2016) Comprehensive investigations of biobutanol production by a non-acetone and 1,3-propanediol generating Clostridium strain from glycerol and polysaccharides. Biotechnol Biofuels 9(220). https://doi.org/10.1186/s13068-016-0641-8
Funding
This study was conducted within project funded by National Academy of Sciences of Ukraine No. 0120U101706 and project funded by Ministry of Education and Science of Ukraine No. 0120U102083. This work also was supported by the Ministry of Education and Science of Ukraine in the project “Cavitation processing of lignocellulosic raw materials in the production of second-generation biofuels” (financed by the European Union’s external aid instrument to fulfill Ukraine’s commitments in the European Union’s Framework Program for Scientific Research and Innovation “Horizon 2020”).
Author information
Authors and Affiliations
Contributions
OT and SS conceived and designed research. OT and MS conducted experiments. VB contributed new reagents or analytical tools. MS and OB analyzed data. LZ performed 16S rRNA gene analysis and protein sequences comparative analysis. LZ and SS wrote the draft. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
This article does not contain studies with human participants or animals performed by any of the authors.
Competing interests
The authors declare no competing interests.
Additional information
Communicated by: Agnieszka Szalewska-Palasz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tigunova, O., Samborskyy, M., Bratishko, V. et al. Main genome characteristics of butanol-producing Clostridium sp. UCM В-7570 strain. J Appl Genetics 64, 559–567 (2023). https://doi.org/10.1007/s13353-023-00766-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-023-00766-8