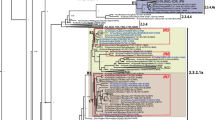
Abstract
Dengue virus (DENV) is the mosquito borne virus which causes Dengue Haemorrhagic Fever and Dengue Shock Syndrome. It consists of four distinct serotypes (DENV 1–4). DENV 1, 3 and 4 were classified into five genotypes (GI–GV), where as DENV-2 belongs to American and Cosmopolitan genotypes. Dengue virus is most prevalent in south and Southeast Asia including India. This study was initiated to study the genetic diversity and evolution among the Dengue isolates in India. Pairwise comparison of amino acid sequences among the serotypes has shown that DENV-3 is having less sequence diversity compared to other serotypes having differences in their amino acid numbers. We have analyzed the 50 Indian strains and 19 of those strains have been identified as recombinant strains by using RDP4 package, which are then excluded for future selection. Episodic positive selection of DENV was obtained using MEME with P value is ≤ 5. Positive selection on several codons was used to correlate the genetic diversity between serotypes. This study clearly established that diversity of amino acids and inter genotypic recombination of strains are the major cause for antigenicity variation and evolution of DENV within India.

Similar content being viewed by others
References
Chen R, Vasilakis N. Dengue—Quo tu et quo vadis. Viruses. 2011;3:1562–608.
Halstead SB. Pathogenesis of dengue: challenges to molecular biology. Science. 1988;239:476–81.
Gubler DJ. Dengue and dengue hemorrhagic fever. Clin Microbiol Rev. 1998;11:480–96.
Waman PV, Mohan MK, Urmila KK. Genetic diversity and evolution of dengue virus serotype 3: a comparative genomics study. Elsevier. 2017;49:234–40.
Amarilla AA, et al. Genetic diversity of the E protein of dengue type 3 virus. Virol J. 2009;6:113. https://doi.org/10.1186/1743-422X-6-113.
Ram S, Khurana S, Kaushal V, Gupta R, Khurana SB. Incidence of dengue fever in relation to climatic factors in Ludhiana, Punjab. Indian J Med Res. 1998;108:128–33.
Singh UB, Maitra A, Broor S, Rai A, Pasha ST, Seth P. Partial nucleotide sequencing and molecular evolution of epidemic causing dengue 2 strains. J Infect Dis. 1999;180:959–65.
Dash PK, Parida MM, Saxena P, Kumar M, Rai A, Pasha ST, Jana AM. Emergence and continued circulation of dengue-2 (genotype IV) virus strains in northern India. J Med Virol. 2004;74:314–22.
Kukreti H, Chaudhary A, Rautela RS, Anand R, Mittal V, Chhabra M, Bhat-tacharya D, Lal S, Rai A. Emergence of an independent lineage ofdengue virus type 1 (DENV-1) and its co-circulation with predominant DENV-3 during the 2006 dengue fever outbreak in Delhi. Int J Infect Dis. 2008;12:542–9.
Amarilla AA, de Almeida FT, Jorge DM, Alfonso HL, de Castro-Jorge LA, Nogueira NA, Figueiredo LT, Aquino VH. Genetic diversity of the E protein of dengue type 3 virus. Virol J. 2009;6:113. https://doi.org/10.1186/1743-422x-6-113.
Annette A, Bennet A, Ajay PJ, Rajendra KB, Suman R, Vinod J. First study of complete genome of Dengue-3 virus from Rajasthan, India: genomic characterization, amino acid variations and phylogenetic analysis. Virol Rep. 2016;6:32–40.
Benson DA, et al. GenBank. Nucleic Acids Res. 2013;41(Database issue):D36–42.
Kolekar P, Kale M, Kulkarni-Kale U. Alignment-free distance measure based on return time distribution for sequence analysis: applications to clustering, molecular phylogeny and subtyping. Mol Phylogenet Evol. 2012;65:510–22.
Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucl Acids Res. 2004;32:1792–7.
Boni MF, Posada D, Feldman MW. An exact nonparametric method for inferring mosaic structure in sequence triplets. Genetics. 2007;176:1035–47.
Martin D, Posada D, Crandall K, Williamson C. A modified bootscan algorithm for automated identification of recombinant sequences and recombination breakpoints. AIDS Res Hum Retrovir. 2005;21:98–102.
Posada D, Crandall KA. Evaluation of methods for detecting recombination from DNA sequences: computer simulations. Proc Natl Acad Sci. 2001;98:13757–62.
Gibbs MJ, Armstrong JS, Gibbs AJ. Sister-scanning: a Monte Carlo procedure for assessing signals in recombinant sequences. Bioinformatics. 2000;16:573–82.
Padidam M, Sawyer S, Fauquet CM. Possible emergence of new geminiviruses by frequent recombination. Virology. 1999;265:218–25.
Smith JM. Analyzing the mosaic structure of genes. J Mol Evol. 1992;34:126–9.
Delport W, Poon AFY, Frost SDW, Pond SL. Datamonkey 2010: a suite of phylogenetic analysis tools for evolutionary biology. Bioinformatics. 2010;26(19):2455–7.
Murrell B, Wertheim JO, Moola S, Weighill T, Scheffler K, Pond SLK. Detecting individual sites subject to episodic diversifying selection. PLoS Genet. 2012;8:e1002764. https://doi.org/10.1371/journal.pgen.1002764.
Brooks AJ, Johansson M, John AV, Xu Y, Jans DA, Vasudevan SG. The interdomain region of dengue NS5 protein that binds to the viral helicase NS3contains independently functional importin beta 1 and importin alpha/beta-recognized nuclear localization signals. J Biol Chem. 2002;277:36399–407.
Dash PK, Sharma S, Soni M, Agarwal A, Parida M, Rao PVL. Complete genome sequencing and evolutionary analysis of Indian isolates of dengue 2virus. Biochem Biophys Res Commun. 2013;436:478–85.
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013;30:2725–9.
Holmes E, Twiddy S. The origin, emergence and evolutionary genetics of dengue virus. Infect Genet Evol. 2003;3:19–28.
Kurane I. Dengue hemorrhagic fever with special emphasis on immunopathogenesis. Comp Immunol Microbiol Infect Dis. 2007;30:329–40.
Matsui K, et al. Characterization of dengue complex-reactive epitopes on dengue 3 virus envelope protein domain III. Virology. 2009;384(1):16–20.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Rexliene, J., Sridhar, J. Genetic diversity and evolutionary dynamics of dengue isolates from India. VirusDis. 30, 354–359 (2019). https://doi.org/10.1007/s13337-019-00538-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-019-00538-1