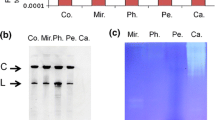
Abstract
Most of the viral diseases of plants are caused by RNA viruses which drastically reduce crop yield. In order to generate resistance against RNA viruses infecting plants, we isolated the dicer 1 protein (CaDcr1), a member of RNAse III family (enzyme that cleaves double stranded RNA) from an opportunistic fungus Candida albicans. In vitro analysis revealed that the CaDcr1 cleaved dsRNA of the coat protein gene of cucumber mosaic virus (genus Cucumovirus, family Bromoviridae). Furthermore, we developed transgenic tobacco plants (Nicotiana tabacum cv. Xanthi) over-expressing expressing CaDcr1 by Agrobacterium mediated transformation. Transgenic tobacco lines were able to suppress infection of an Indian isolate of potato virus X (genus Potexvirus, family Alphaflexiviridae). The present study demonstrates that CaDcr1 can cleave double stranded replicative intermediate and provide tolerance to plant against RNA viruses.






Similar content being viewed by others
References
Aravind L, Koonin EV. A natural classification of ribonucleases. Methods Enzymol. 2001;341:3–28.
Bernsteina DA, Vyasa VK, Weinberga DE, Drinnenberga IA, Bartela DP, Finka GR. Candida albicans Dicer (CaDcr1) is required for efficient ribosomal and spliceosomal RNA maturation. PNAS USA. 2012;109:523–8.
Braun BR, van het Hoog M, d’Enfert C, Martchenko M, Dungan J, Kuo A, Inglis DO, Uhl MA, Hogues H, Berriman M, Lorenz M, Levitin A, Oberholzer U, Bachewich C, Harcus D, Marcil A, Dignard D, Iouk T, Rosa Z, Frangeul L, Tekaia F, Rutherford K, Wang E, Munro CA, Bates S, Gow NA, Hoyer LL, Köhler G, Morschhäuser J, Newport G, Znaidi S, Raymond M, Turcotte B, Sherlock G, Costanzo M, Ihmels J, Berman J, Sanglard D, Agabian N, Mitchell AP, Johnson AD, Whiteway M, Nantel A. A human-curated annotation of the Candida albicans genome. PLoS Genet. 2005;1(1):36–57.
Chanfreau G, Rotondo G, Legrain P, Jacquier A. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 1998;17:3726–37.
Conrad C, Rauhut R. Ribonuclease III: new sense from nuisance. Int J Biochem Cell Biol. 2002;34:116–29.
Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Mol Biol Rep. 1983;1:19–21.
Ding SW, Voinnet O. Antiviral immunity directed by small RNAs. Cell. 2007;130:413–26.
Drinnenberg IA, Weinberg DE, Xie KT, Mower JP, Wolfe KH, Fink GR, Bartel DP. RNAi in budding yeast. Science. 2009;326:544–50.
Elela SA, Igel H, Ares M. RNase III cleaves eukaryotic preribosomal RNA at a U3 snoRNP-dependent site. Cell. 1996;85:115–24.
Filippov V, Solovyev V, Filippova M, Gill SS. A novel type of RNase III family proteins in eukaryotes. Gene. 2000;245:213–21.
Geetanjali AS, Kumar R, Srivastava PS, Mandal B. Biological and molecular characterization of two distinct tomato strains of Cucumber mosaic virus based on complete RNA-3 genome and subgroup specific diagnosis. Indian J Virol. 2011;22:117–26.
Giuseppe R, David F. Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res. 1996;24(12):2377–86.
Horsch RB, Rogers SG, Fraley RT. Transgenic plants. Cold Spring Harb Symp Quant Biol. 1985;50:433–7.
Jacobsen SE, Running MP, Meyerowitz EM. Disruption of an RNA helicase/RNAse III gene in Arabidopsis causes unregulated cell division in floral meristems. Development. 1999;126:5231–43.
Iino Y, Sugimoto A, Yamamoto M. Schizosachraomyces pombe pac1+ , whose overexpression inhibits sexual development, encodes a ribonuclease III-like RNase. EMBO J. 1991;10:221–6.
Kiyota E, Okada R, Kondo N, Hiraguri A, Moriyama H, Fukuhara T. Arabidopsis RNase III-like protein, AtRTL2, cleaves double-stranded RNA in vitro. J Plant Res. 2001;124:405–14.
Kumari P, Singh AK, Sharma VK, Chattopadhyay B, Chakraborty S. A novel recombinant tomato-infecting begomovirus capable of trans-complementing heterologous DNA-B components. Arch Virol. 2001;156:769–83.
Lamontagne B, Trembley A, Elela SA. The N-Terminal domain that distinguishes yeast from bacterial RNase III contains dimerization signal required for efficient double-stranded RNA cleavage. Mol Cell Biol. 2000;20:1104–15.
Lamontagne B, Elela SA. Purification and characterization of Saccharomyces cerevisiae Rnt1p nuclease. Methods Enzymol. 2001;342:159–67.
Langenberg WG, Zhang L, Court D, Giunchedi L, Mirtra A. Transgenic tobacco plants expressing the bacterial rnc gene resist virus infection. Mol Breed. 1997;3:391–9.
Li H, Nicholson AW. Defining the enzyme binding domain of a ribonuclease III processing signal. Ethylation interference and hydroxyl radical foot printing using catalytically inactive RNase III mutants. EMBO J. 1996;15:1421–33.
MacRae IJ, Doudna JA. Ribonuclease revisited: structural insights into ribonuclease III family enzymes. Curr Opin Struct Biol. 2007;17:138–45.
MacRae IJ, Zhou K, Doudna JA. Structural determinants of RNA recognition and cleavage by Dicer. Nat Struct Mol Biol. 2007;14:934–40.
Malone CD, Hannon GJ. Small RNAs as guardians of the genome. Cell. 2009;136:656–68.
Moore JT, Uppal A, Maley F, Maley GF. Overcoming inclusion body formation in a high-level expression system. Protein Express Purif. 1993;4:160–3.
Nicholson AW. Structure, reactivity, and biology of double-stranded RNA. Prog Nucleic Acid Res Mol Biol. 1996;52:1–65.
Nicholson AW. Function, mechanism and regulation of bacterial ribonucleases. FEMS Microbiol Rev. 1999;23:371–90.
Robaglia C, Tepfer M. Using nonviral genes to engineer virus resistance in plants. Biotechnol Annu Rev. 1996;2:185–204.
Robertson HD, Webster RE, Zinder ND. Purification and properties of ribonuclease III from Escherichia coli. J Biol Chem. 1968;243(1):82–91.
Rotondo G, Frendewey D. Purification and characterization of the Pac1 ribonuclease of Schizosaccharomyces pombe. Nucleic Acids Res. 1996;24:2377–86.
Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3rd ed. Cold Spring Harbor: Cold Spring Harbor Laboratory; 2001.
Sano T, Nagayama A, Ogawa T, Ishida I, Okada Y. Transgenic potato expressing a double-stranded RNA-specific ribonuclease is resistant to potato spindle tuber viroid. Nat Biotechnol. 1997;15:1290–4.
Sharma K, Misra RS. Molecular approaches towards analyzing the viruses infecting maize (Zea mays L.). J Gen Mol Virol. 2011;3(1):1–17.
Varma A, Malathi VG. Emerging geminivirus problems: a serious threat to crop production. Ann Appl Biol. 2003;142:145–64.
Watanabe Y, Ogawa T, Takahashi H, Ishida I, Takeuchi Y, Yamamoto M, Okada Y. Resistance against multiple plant viruses in plants mediated by a double-stranded RNA-specific ribonuclease. FEBS Lett. 1995;372:165–8.
Weinberg DE, Nakanishi K, Patel DJ, Bartel DP. The inside-out mechanism of Dicers from budding yeasts. Cell. 2011;146:262–76.
Xu HP, Riggs M, Rodgers L, Wigler MA. A gene from S. pombe with homology to E. coli RNAse III blocks conjugation and sporulation when overexpressed in wild type cells. Nucleic Acids Res. 1990;18(17):5304.
Zamore PD, Haley B. Ribo-gnome: the big world of small RNAs. Science. 2005;309:1519–24.
Acknowledgements
Financial support from the Council for Scientific and Industrial Research, Govt. of India vide Grant No. 38(1300)/11/EMR-11 to SC is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing financial interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
Confirmation of the recombinant CaDcr1 protein by MALDI analysis (TIFF 548 kb)
Fig. S2
Secondary structure of the coat protein transcripts of CMV and free energy value using GeneBee software.(a) Secondary structure of transcript of CMV-AUR(-133.6 kcal/mol free energy) (b) Secondary structure of transcript of CMV-PUNE (-147.0 kcal/mol free energy) (TIFF 2794 kb)
Fig. S3
Hyperchromicity Graph for determination melting point of secondary structure of the coat protein transcripts of CMV using Poland Software.(a) AUR RNA (100 °C) and (b) PUNE RNA (100 °C) (TIFF 83 kb)
Fig. S4
Prediction of allergenicity of CaDcr1. Non-Allergic nature of CaDcr1 was established using the Structural database of Allergen Protein (SDAP) software. The allergenicity value ranges between 3 to 11% that are much lower than the required threshold value (35%) as set for known allergens (indicated as Red horizontal line) (TIFF 215 kb)
Rights and permissions
About this article
Cite this article
Alam, C.M., Jain, G., Kausar, A. et al. Dicer 1 of Candida albicans cleaves plant viral dsRNA in vitro and provides tolerance in plants against virus infection. VirusDis. 30, 237–244 (2019). https://doi.org/10.1007/s13337-019-00520-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13337-019-00520-x