Abstract
Background
The demand for physiologically based pharmacokinetic (PBPK) model is increasing currently. New drug application (NDA) of many compounds is submitted with PBPK models for efficient drug development. Tissue-to-plasma partition coefficient (Kp) is a key parameter for the PBPK model to describe differential equations. However, it is difficult to obtain the Kp value experimentally because the measurement of drug concentration in the tissue is much harder than that in plasma.
Objective
Instead of experiments, many researchers have sought in silico methods. Today, most of the models for Kp prediction are using in vitro and in vivo parameters as explanatory variables. We thought of physicochemical descriptors that could improve the predictability. Therefore, we aimed to develop the two-dimensional quantitative structure-activity relationship (2D-QSAR) model for Kp using physicochemical descriptors instead of in vivo experimental data as explanatory variables.
Methods
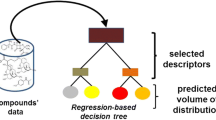
We compared our model with the conventional models using 20-fold cross-validation according to the published method (Yun et al. J Pharmacokinet Pharmacodyn 41:1–14, 2014). We used random forest algorithm, which is known to be one of the best predictors for the 2D-QSAR model. Finally, we combined minimum in vitro experimental values and physiochemical descriptors. Thus, the prediction method for Kp value using a few in vitro parameters and physicochemical descriptors was developed; this is a multimodal model.
Results
Its accuracy was found to be superior to that of the conventional models. Results of this research suggest that multimodality is useful for the 2D-QSAR model [RMSE and % of two-fold error: 0.66 and 42.2% (Berezohkovsky), 0.52 and 52.2% (Rodgers), 0.65 and 34.6% (Schmitt), 0.44 and 61.1% (published model), 0.41 and 62.1% (traditional model), 0.39 and 64.5% (multimodal model)].
Conclusion
We could develop a 2D-QSAR model for Kp value with the highest accuracy using a few in vitro experimental data and physicochemical descriptors.




Similar content being viewed by others
References
PMDA. Guidelines for Non-Clinical Pharmacokinetic Studies. No 469. 1998.
Model-Informed Drug Development Pilot Program. https://www.fda.gov/drugs/development-resources/model-informed-drug-development-paired-meeting-program. Accessed 8 May 2023.
US Department of Health and Human Services Food and Drug Administration Center for Drug Evaluation and Research (CDER). Physiologically Based Pharmacokinetic Analyses—Format and Content Guidance for Industry. 2019.
EMA. Reporting of physiologically based pharmacokinetic (PBPK) modelling and simulation. European Medicines Agency. 2018. https://www.ema.europa.eu/en/reporting-physiologically-based-pharmacokinetic-pbpk-modelling-simulation-scientific-guideline. Accessed 10 Apr 2023.
Shebley M, Sandhu P, Emami Riedmaier A, Jamei M, Narayanan R, Patel A, et al. Physiologically based pharmacokinetic model qualification and reporting procedures for regulatory submissions: a consortium perspective. Clin Pharmacol Ther. 2018;104(1):88–110.
Perry C, Davis G, Conner TM, Zhang T. Utilization of physiologically based pharmacokinetic modeling in clinical pharmacology and therapeutics: an overview. Curr Pharmacol reports. 2020;6(3):71–84.
Jones H, Rowland-Yeo K. Basic concepts in physiologically based pharmacokinetic modeling in drug discovery and development. CPT Pharmacomet Syst Pharmacol. 2013;2(8): e63.
Ho S. Challenges of atypical matrix effects in tissue. Bioanalysis. 2013;5(19):2333–5.
Poulin P, Theil FP. A priori prediction of tissue:plasma partition coefficients of drugs to facilitate the use of physiologically-based pharmacokinetic models in drug discovery. J Pharm Sci. 2000;89(1):16–35.
Poulin P, Schoenlein K, Theil FP. Prediction of adipose tissue: plasma partition coefficients for structurally unrelated drugs. J Pharm Sci. 2001;90(4):436–47.
Poulin P, Krishnan K. A biologically-based algorithm for predicting human tissue: blood partition coefficients of organic chemicals. Hum Exp Toxicol. 1995;14(3):273–80.
Berezhkovskiy LM. Volume of distribution at steady state for a linear pharmacokinetic system with peripheral elimination. J Pharm Sci. 2004;93(6):1628–40.
Rodgers T, Rowland M. Physiologically based pharmacokinetic modelling 2: predicting the tissue distribution of acids, very weak bases, neutrals and zwitterions. J Pharm Sci. 2006;95(6):1238–57.
Schmitt W. General approach for the calculation of tissue to plasma partition coefficients. Toxicol In Vitro. 2008;22(2):457–67.
Björkman S. Reduction and lumping of physiologically based pharmacokinetic models: prediction of the disposition of fentanyl and pethidine in humans by successively simplified models. J Pharmacokinet Pharmacodyn. 2003;30(4):285–307.
Lusher SJ, McGuire R, van Schaik RC, Nicholson CD, de Vlieg J. Data-driven medicinal chemistry in the era of big data. Drug Discov Today. 2014;19(7):859–68.
Yun YE, Cotton CA, Edginton AN. Development of a decision tree to classify the most accurate tissue-specific tissue to plasma partition coefficient algorithm for a given compound. J Pharmacokinet Pharmacodyn. 2014;41(1):1–14.
Russell WMS, Burch RL. The principles of humane experimental technique. Wheathampstead: Universities Federation for Animal Welfare; 1959.
Lo Y-C, Rensi SE, Torng W, Altman RB. Machine learning in chemoinformatics and drug discovery. Drug Discov Today. 2018;23(8):1538–46.
Srivastava N, Salakhutdinov RR. Multimodal learning with deep Boltzmann machines. J Mach Learn Res. 2014;15:2949–80.
Hofmann-Apitius M, Ball G, Gebel S, Bagewadi S, de Bono B, Schneider R, et al. Bioinformatics mining and modeling methods for the identification of disease mechanisms in neurodegenerative disorders. Int J Mol Sci. 2015;16(12):29179–206.
Goh GB, Sakloth K, Siegel C, Vishnu A, Pfaendtner J. Multimodal Deep Neural Networks using Both Engineered and Learned Representations for Biodegradability Prediction. 2018 13; http://arxiv.org/abs/1808.04456
Wajima T, Fukumura K, Yano Y, Oguma T. Prediction of human clearance from animal data and molecular structural parameters using multivariate regression analysis. J Pharm Sci. 2002;91(12):2489–99.
Iwata H, Matsuo T, Mamada H, Motomura T, Matsushita M, Fujiwara T, et al. Prediction of total drug clearance in humans using animal data: proposal of a multimodal learning method based on deep learning. J Pharm Sci. 2021;110(4):1834–41.
Schneckener S, Grimbs S, Hey J, Menz S, Osmers M, Schaper S, Hillisch A, Göller AH. Prediction of oral bioavailability in rats: transferring insights from in vitro correlations to (Deep) machine learning models using in silico model outputs and chemical structure parameters. J Chem Inf Model. 2019;59(11):4893–905.
Kosugi Y, Hosea N. Prediction of oral pharmacokinetics using a combination of in silico descriptors and in vitro ADME properties. Mol Pharm. 2021;18(3):1071–9.
Obrezanova O, Martinsson A, Whitehead T, Mahmoud S, Bender A, Miljković F, Grabowski P, Irwin B, Oprisiu I, Conduit G, Segall M, Smith GF, Williamson B, Winiwarter S, Greene N. Prediction of in vivo pharmacokinetic parameters and time-exposure curves in rats using machine learning from the chemical structure. Mol Pharm. 2022;19(5):1488–504.
Iwata H, Matsuo T, Mamada H, Motomura T, Matsushita M, Fujiwara T, Maeda K, Handa K. Predicting total drug clearance and volumes of distribution using the machine learning-mediated multimodal method through the imputation of various nonclinical data. J Chem Inf Model. 2022;62(17):4057–65.
PubChem: https://pubchem.ncbi.nlm.nih.gov. Accessed 10 Apr 2023.
Ligprep: https://www.schrodinger.com/products/ligprep. Accessed 10 Apr 2023.
ADMET Predictor software: https://www.simulations-plus.com/software/gastroplus/discovery-pbpk. Accessed 10 Apr 2023.
Clark RD, Morris DN, Chinigo G, Lawless MS, Prudhomme J, et al. Design and tests of prospective property predictions for novel antimalarial 2-aminopropylaminoquinolones. J Comput Aided Mol Des. 2020;34(11):1117–32.
Clark RD. Predicting mammalian metabolism and toxicity of pesticides in silico. Pest Manag Sci. 2018;74(9):1992–2003.
Daga PR, Bolger MB, Haworth IS, Clark RD, Martin EJ. Physiologically Based Pharmacokinetic Modeling in Lead Optimization. 2. Rational Bioavailability Design by Global Sensitivity Analysis To Identify Properties Affecting Bioavailability. Mol Pharm. 2018;15(3):831–39.
Pipeline Pilot 2016: https://www.3ds.com/products-services/biovia/products/data-science/pipeline-pilot. Accessed 10 Apr 2023.
Breiman L. Random forests. Mach Learn. 2001;45:5–32.
De P, Kar S, Ambure P, Roy K. Prediction reliability of QSAR models: an overview of various validation tools. Arch Toxicol. 2022;96(5):1279–95. https://doi.org/10.1007/s00204-022-03252-y. (Epub 2022 Mar 10 PMID: 35267067).
Eertink JJ, Heymans MW, Zwezerijnen GJC, Zijlstra JM, de Vet HCW, Boellaard R. External validation: a simulation study to compare cross-validation versus holdout or external testing to assess the performance of clinical prediction models using PET data from DLBCL patients. EJNMMI Res. 2022;12(1):58.
Isbell J, Yuan D, Torrao L, Gatlik E, Hoffmann L, Wipfli P. Plasma protein binding of highly bound drugs determined with equilibrium gel filtration of nonradiolabeled compounds and LC-MS/MS detection. J Pharm Sci. 2019;108(2):1053–60.
Chen C, Zhou H, Guan C, Zhang H, Li Y, Jiang X, Dong Z, Tao Y, Du J, Wang S, Zhang T, Du N, Guo J, Wu Y, Song Z, Luan H, Wang Y, Du H, Zhang S, Li C, Chang H, Wang T. Applicability of free drug hypothesis to drugs with good membrane permeability that are not efflux transporter substrates: a microdialysis study in rats. Pharmacol Res Perspect. 2020;8(2): e00575.
GUIDANCE DOCUMENT: In Vitro Drug Interaction Studies—Cytochrome P450 Enzyme- and Transporter-Mediated Drug Interactions Guidance for Industry. https://www.fda.gov/regulatory-information/search-fda-guidance-documents/in-vitro-drug-interaction-studies-cytochrome-p450-enzyme-and-transporter-mediated-drug-interactions. Accessed 5 Apr 2023.
Guideline on the investigation of drug interactions. https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-investigation-drug-interactions-revision-1_en.pdf. Accessed 5 Apr 2023.
Riccardi K, Cawley S, Yates PD, Chang C, Funk C, Niosi M, Lin J, Di L. Plasma protein binding of challenging compounds. J Pharm Sci. 2015;104(8):2627–36.
Tietz S, Engelhardt B. Brain barriers: crosstalk between complex tight junctions and adherens junctions. J Cell Biol. 2015;209(4):493–506.
Pardridge WM. Blood-brain barrier biology and methodology. J Neurovirol. 1999;5(6):556–69.
Ayloo S, Gu C. Transcytosis at the blood-brain barrier. Curr Opin Neurobiol. 2019;57:32–8.
César-Razquin A, Snijder B, Frappier-Brinton T, Isserlin R, Gyimesi G, Bai X, et al. A call for systematic research on solute carriers. Cell. 2015;162(3):478–87.
Mikkaichi T, Nakai D, Yoshigae Y, Imaoka T, Okudaira N, Izumi T. Liver-selective distribution in rats supports the importance of active uptake into the liver via organic anion transporting polypeptides (OATPs) in humans. Drug Metab Pharmacokinet. 2015;30(5):334–40.
Kawai R, Lemaire M, Steimer JL, Bruelisauer A, Niederberger W, Rowland M. Physiologically based pharmacokinetic study on a cyclosporin derivative, SDZ IMM 125. J Pharmacokinet Biopharm. 1994;22(5):327–65.
Orozco CC, Atkinson K, Ryu S, Chang G, Keefer C, Lin J, et al. Structural attributes influencing unbound tissue distribution. Eur J Med Chem. 2020;185: 111813.
Tiwari R, Jaimini M, Mohan S, Sharma S. Transdermal drug delivery system: a review. Int J Ther Appl. 2013;14:22–8.
Baba H, Ueno Y, Hashida M, Yamashita F. Quantitative prediction of ionization effect on human skin permeability. Int J Pharm. 2017;522(1–2):222–33.
Baba H, Takahara J, Mamitsuka H. In silico predictions of human skin permeability using nonlinear quantitative structure-property relationship models. Pharm Res. 2015;32(7):2360–71.
Baba H, Takahara J, Yamashita F, Hashida M. Modeling and prediction of solvent effect on human skin permeability using support vector regression and random forest. Pharm Res. 2015;32(11):3604–17.
Sahigara F, Mansouri K, Ballabio D, Mauri A, Consonni V, Todeschini R. Comparison of different approaches to define the applicability domain of QSAR models. Molecules. 2012;17(5):4791–810.
Acknowledgments
We thank the researchers at the Teijin Institute for Bio-Medical Research who encouraged us to carry out this study.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethical Approval
Not applicable.
Competing interests
The authors have no competing interests to disclose.
Authors' Contributions
Koichi Handa wrote the manuscript. Seishiro Sakamoto also wrote the manuscript. Michiharu Kageyama made the figures and tables. Takeshi Iijima reviewed the manuscript.
Funding
The author(s) reported there is no funding associated with the work featured in this article.
Availability of Data and Materials
All the data can be obtained from the corresponding author according to requests.
Code Availability
All of the software used in this study is freely available, as are the codes, through the software package described in the manuscript.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Handa, K., Sakamoto, S., Kageyama, M. et al. Development of a 2D-QSAR Model for Tissue-to-Plasma Partition Coefficient Value with High Accuracy Using Machine Learning Method, Minimum Required Experimental Values, and Physicochemical Descriptors. Eur J Drug Metab Pharmacokinet 48, 341–352 (2023). https://doi.org/10.1007/s13318-023-00832-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13318-023-00832-w