Abstract
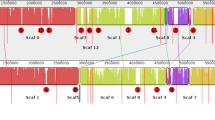
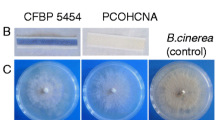
Pseudomonas protegens (formerly Pseudomonas fluorescens) FD6, which was isolated from the canola rhizosphere, is a biocontrol agent that protects against the fungal pathogens Botrytis cinerea and Monilinia fructicola. The complete sequence of the 6.7-Mb Pseudomonas protegens FD6 genome was determined. Genomic analysis of strain FD6 led to the identification of twelve putative gene clusters of secondary metabolites, including the antibiotics 2,4-diacetylphloroglucinol (2,4-DAPG), pyoluteorin (PLT) and pyrrolnitrin (PRN). To assess the role of 2,4-DAPG and PLT in the biocontrol activity of P. protegens FD6, two mutants of P. protegens FD6, including strains ΔphlC and ΔpltD, were constructed using homologous recombination technology. The results showed that no significant differences were observed in the antagonistic action of the ΔphlC mutant compared with wild-type FD6. However, the ΔpltD mutant no longer exhibited antifungal activity. HPLC analysis indicated that ΔphlC mutant could not biosynthesize 2,4-DAPG, whereas PLT production in the ΔphlC mutant was significantly increased by 67-fold compared with wild-type FD6. Biosynthesis of PLT in the ΔpltD mutant was not detected, whereas the level of 2,4-DAPG was vastly decreased over sixteenfold compared with strain FD6. The major antagonistic activity produced by P. protegens FD6 corresponded to 2,4-diacetylphloroglucinol and pyoluteorin. There is an inverse interaction between 2,4-DAPG and PLT.






Similar content being viewed by others
References
Achkar J, Xian M, Zhao H, Frost JW (2005) Biosynthesis of phloroglucinol. J Am Chem Soc 127:5332–5333
Bangera MG, Thomashow LS (1999) Identification and characterization of a gene cluster for synthesis of the polyketide antibiotic 2,4-diacetylphloroglucinol from Pseudomonas fluorescens Q2-87. J Bacteriol 181:3155–3163
Castric KF, Castric PA (1983) Method for rapid detection of cyanogenic bacteria. Appl Environ Microbiol 45:701–702
Chang L, Li Q, Tong YH, Xu JY, Zhang QX (2011) Identification of the biocontrol bacterial strain FD6 and antimicrobial study of this bacterium against tomato grey mould pathogen Botrytis cinerea. Acta Phytophylacica Sin 38:487–492
Chang L, Xiao Q, Tong YH, Xu JY, Zhang QX (2014) Functional analysis of the gacS gene in a tomato grey mould suppressive bacterium Pseudomonas fluorescens FD6. Acta Horticulturae Sin 41:681–686
Choi O, Kim J, Kim JG, Jeong Y, Moon JS, Park CS, Hwang I (2008) Pyrroloquinline quinone is a plant growth promotion factor produced by Pseudomomas fluorescens B16. Plant Physiol 146:657–668
Costa R, Aarle IMV, Mendes R, Elsas JDV (2009) Genomics of pyrrolnitrin biosynthetic loci: evidence for conservation and whole-operon mobility within gram-negative bacteria. Environ Microbiol 11:159–175
Dispirito AA, Zahn JA, Graham DW, Kim HJ, Michail A, Cynthia L (2007) Methanobactin: a copper binding compound having antibiotic and antioxidant activity isolated from methanotrophic bacteria. US patent no. 7,199,099 B2
Dorrestein PC, Yeh E, Garneau-Tsodikova S, Kelleher NL, Walsh CT (2005) Dichlorination of a pyrrolyl-S-carrier protein by FADH2-dependent halogenase PltA during pyoluteorin biosynthesis. PNAS 102(39):13843–13848
Duan J, Jiang W, Cheng ZY, Heikkila JJ, Glick BR (2013) The complete genome sequence of the plant growth-promoting bacterium Pseudomonas sp. UW4. PLoS One 8:e58640
Gross H, Loper JE (2007) Genomic analysis of antifungal metabolite production by Pseudomonas fluorescens Pf-5. Eur J Plant Pathol 119:265–278
Gross H, Loper JE (2009) Genomics of secondary metabolite production by Pseudomonas spp. Nat Prod Rep 26:1408–1446
Haas D, Defago G (2005) Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat Rev Microbiol 3:307–319
Hoang TT, Karkhoff-Schweizer RAR, Kutchma AJ, Schweizer HP (1998) A broad-host-range Flp-FRT recombination system for site-specific excision of chromosomally-located DNA sequences: application for isolation of unmarked Pseudomonas aeruginosa mutants. Gene 212:77–86
Howell CR, Stipanovic RD (1980) Suppression of Pythium ultimum-induced damping-off of cotton seedlings by Pseudomonas fluorescens and its antibiotic, pyoluteorin. Phytopathology 70:712–715
Hu HB, Xu YQ, Chen F, Zhang XH, Hur BK (2005) Isolation and characterization of a new fluorescent Pseudomonas strain that produces both phenazine 1-carboxylic acid and pyoluteorin. J Microbiol Biotechnol 15(1):86–90
Kenney GE, Dassama LMK, Pandelia ME, Gizzi AS, Martinie RJ, Gao P, DeHart CJ, Schachner LF, Skinner OS, Ro SY, Zhu X, Sadek M, Thomas PM, Almo SC, Bollinger JM, Krebs C, Kelleher NL, Rosenzweig AC (2018) The biosynthesis of methanobactin. Science 359:1411–1416
Kidarsa TA, Goebel NC, Zabriskie TM, Loper JE (2011) Phloroglucinol mediates cross-talk between the pyoluteorin and 2,4-diacetylphloroglucinol biosynthetic pathways in Pseudomonas fluorescens Pf-5. Mol Microbiol 81:395–414
Kraus J, Loper JE (1995) Characterization of a genomic region required for production of the antibiotic pyoluteorin by the biological control agent Pseudomonas fluorescens Pf-5. Appl Environ Microbiol 61:849–854
Lagesen K, Hallin P, Rødland EA, Staerfeldt HH, Ussery DW (2007) RNAmmer: consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res 35:3100–3108
Laslett D, Canback B (2004) ARAGORN, a program to detect tRNA genes and tmRNA genes in nucleotide sequences. Nucleic Acids Res 32:11–16
Li SN, Li K, Wang SY, Wang GH, Huang XQ, Xu YQ (2012) Identification and expression optimization of the genes encoding rate-limiting enzymes in pyoluteorin biosynthesis of Pseudomonas sp. M18. Microbiol Chin 39(3):300–308
Liu H, Dong D, Peng H, Zhang X, Xu Y (2006) Genetic diversity of phenazine- and pyoluteorin-producing pseudomonads isolated from green pepper rhizosphere. Arch Microbiol 185:91–98
Liu XG, Bimerew M, Ma YX, Muller H, Ovadis M, Eberl L (2010) Quorum-sensing signaling is required for production of the antibiotic pyrrolnitrin in a rhizospheric biocontrol strain of Serratia plymuthica. FEMS Microbiol Lett 270:299–305
Liu YZ, Lu SE, Sonya MB, Qiao JQ, Du Y (2015) Cloning the genes of Pseudomonas chlororaphis YL-1 dedicated to antibacterial activities against microbial phytopathogens. Acta Phytopathologica Sin 3:307–316
Loper JE, Henkels MD, Shaffer BT, Valeriote FA, Gross H (2008) Isolation and identifification of rhizoxin analogs from Pseudomonas fluorescens Pf-5 by using a genomic mining strategy. Appl Environ Microbiol 74(10):3085–3093
Loper JE, Hassan KA, Mavrodi DV, Davis EW, Lim CK, Shaffer BT, Elbourne LD, Stockwell VO, Hartney SL, Breakwell K, Henkels MD, Tetu SJ, Rangel LL, Kidarsa TA, Wilson NL, van de Mortel JE, Song CX, Blumhagen R, Radune D, Hostetler JB, Brinkac LM, Durkin AS, Kluepfel DA, Wechter WP, Anderson AJ, Kim YC, Pierson LS, Pierson EA, Lindow SE, Kobayashi DY, Raaijmakers JM, Weller DM, Thomashow LS, Allen AE, Paulsen LT (2012) Comparative genomics of plant-associated Pseudomonas spp.: insights into diversity and inheritance of traits involved in multitrophic interactions. PLoS Genet 8:e1002784
Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25:955–964
Ma Z, Geudens N, Kieu NP, Sinnaeve D, Ongena M, Martins JC, Höfte M (2016) Biosynthesis, chemical structure, and structure-activity relationship of orfamide lipopeptides produced by Pseudomonas protegens and related species. Front Microbiol 7:382
Nawrocki EP, Eddy SR (2013) Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29:2933–2935
Nawrocki EP, Burge SW, Bateman A, Daub J, Eberhardt RY, Eddy SR, Floden EW, Gardner PP, Jones TA, Tate J, Finn RD (2015) Rfam 12.0: updates to the RNA families database. Nucleic Acids Res 43:D130–D137
Nowak-Thompson B, Gould SJ, Kraus J, Loper JE (1994) Production of 2,4-diacetylphloroglucinol by the biocontrol agent Pseudomonas fluorescens, Pf-5. Can J Microbiol 40:1064–1066
Nowak-Thompson B, Chaney N, Wing JS, Gould SJ, Loper JE (1999) Characterization of the pyoluteorin biosynthetic gene cluster of Pseudomonas fluorescens Pf5. J Bacteriol 181(7):2166–2174
O’Brien PA (2017) Biological control of plant diseases. Australas Plant Pathol 46(4):1–12
Periasamy S, Nair HAS, Lee KWK, Ong J, Goh JQJ, Kjelleberg S, Rice SA (2015) Pseudomonas aeruginosa PAO1 exopolysaccharides are important for mixed species biofilm community development and stress tolerance. Front Microbiol 6:851
Poritsanos N, Selin C, Fernando WGD, Nakkeeran S, Kievit TRD (2006) A GacS deficiency does not affect Pseudomonas chlororaphis PA23 fitness when growing on canola, in aged batch culture or as a biofilm. Can J Microbiol 52:1177–1188
Raaijmakers JM, Mazzola M (2012) Diversity and natural functions of antibiotics produced by beneficial and plant pathogenic bacteria. Annu Rev Phytopathol 50:403–424
Ramette A, Frapolli M, Saux MF-L, Gruffaz C, Meyer JM, Défago G, Sutra L, Moënne-Loccoz Y (2011) Pseudomonas protegens sp. nov., widespread plant-protecting bacteria producing the biocontrol compounds 2,4-diacetylphloroglucinol and pyoluteorin. Syst Appl Microbiol 34:180–188
Reimmann C, Beyeler M, Latifi A, Winteler H, Foglino M, Lazdunski A (1997) The global activator GacA of Pseudomonas aeruginosa PAO positively controls the production of the autoinducer N-butyryl-homoserine lactone and the formation of the virulence factors pyocyanin, cyanide, and lipase. Mol Microbiol 24:309–319
Russell DW, Sambrook J (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor
Ryu CM, Farag MA, Hu CH, Reddy MS, Wei HX, Pare PW, Kloepper JW (2003) Bacterial volatiles promote growth in Arabidopsis. Proc Natl Acad Sci U S A 100:4927–4932
Schwyn B, Neilands JB (1987) Universal chemical assay for the detection and determination of siderophores. Anal Biochem 160:47–56
Shen X, Hu H, Peng H, Wang W, Zhang X (2013) Comparative genomic analysis of four representative plant growth-promoting rhizobacteria in Pseudomonas. BMC Genomics 14:271
Strano CP, Bella P, Licciardello G, Caruso A, Catara V (2017) Role of secondary metabolites in the biocontrol activity of Pseudomonas corrugata and Pseudomonas mediterranea. Eur J Plant Pathol 149:103–115
Wang C, Knill E, Glick BR, Geneviève D (2000) Effect of transferring 1-aminocyclopropane-1-carboxylic acid (Acc) deaminase genes into Pseudomonas fluorescens strain CHA0 and its gacA derivative CHA96 on their growth-promoting and disease-suppressive capacities. Can J Microbiol 46:898–907
Weber T, Blin K, Duddela S, Krug D, Kim HU, Bruccoleri R, Lee SY, Fischbach MA, Müller R, Wohlleben W, Breitling R, Takano E, Medema MH (2015) antiSMASH 3.0-a comprehensive resource for the genome mining of biosynthetic gene clusters. Nucleic Acids Res 43:W237–W243
Wei HL, Zhang LQ (2006) Quorum-sensing system influences root colonization and biological control ability in Pseudomonas fluorescens 2P24. Antonie Van Leeuwenhoek 89:267–280
Wei HL, Zhou HY, Zhang LQ, Wang Y, Tang WH (2004) Experimental evidence on the functional agent of 2,4-diacetylphloroglucinol in biocontrol activity of Pseudomonas fluorescens 2P24. Acta Microbiol Sin 44:663–666
Weller DM (2007) Pseudomonas biocontrol agents of soilborne pathogens: looking back over 30 years. Phytopathology 97:250–256
Yan Q, Philmus B, Chang JH, Loper JE (2017) Novel mechanism of metabolic co-regulation coordinates the biosynthesis of secondary metabolites in Pseudomonas protegens. eLife 6:e22835
Zhang W, Zhao Z, Zhang B, Wu XG, Zhang LQ (2014) Posttranscriptional regulation of 2,4-diacetylphloroglucinol production by GidA and TrmE in Pseudomonas fluorescens 2P24. Appl Environ Microbiol 80:3972–3981
Zhang QX, Xiao Q, Xu JY, Tong YH, Wen J, Chen XJ, Wei LH (2015) Effect of retS gene on antibiotics production in Pseudomonas fluorescens FD6. Microbiol Res 180:23–29
Zhang QX, He LL, Shan HH, Tong YH, Chen XJ, Ji ZL, Liu FQ (2016) Cloning of pyrrolnitrin synthetic gene cluster prn and prnA functional analysis from antagnistic bacteria FD6 against peach brown rot. Acta Horticulturae Sin 8:1473–1481
Zhang QX, Zhang Y, He LL, Ji ZL, Tong YH (2018) Identification of a small antimycotic peptide produced by Bacillus amyloliquefaciens 6256. Pestic Biochem Physiol 150:78–82
Acknowledgements
This study was funded by the National Natural Science Foundation (31772210), the Jiangsu Provincial Key Research and Development Program (BE2017344, BE2018359), the Key Science and Technology Program of Yangzhou City (YZ2018137), and the Shandong Provincial Key Laboratory of Agricultural Microbiology Open Fund (SDKL2017015).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Zhang, Q.X., Kong, X.W., Li, S.Y. et al. Antibiotics of Pseudomonas protegens FD6 are essential for biocontrol activity. Australasian Plant Pathol. 49, 307–317 (2020). https://doi.org/10.1007/s13313-020-00696-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-020-00696-7