Abstract
After the detection of the myrtle rust pathogen, Austropuccinia psidii, in New Zealand, a biosecurity response was initiated, including a wide-spread surveillance programme. Through an intensive public awareness initiative, the general public was highly engaged in reporting myrtle rust infections and added significant value to the surveys by reporting first detections from most of the areas that are now known to be infected. During the first year of the response, Austropuccinia psidii was found in areas that were predicted to be at high infection risk in previous modelling studies. Significant surveillance resources were deployed to different parts of the country and the response surveillance team contributed to most of the new host species finds. Twenty -four species and six hybrids of Myrtaceae have been confirmed to be naturally infected by myrtle rust in New Zealand. Eleven of these are new host records globally and three were previously recorded only as experimental hosts.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The causal agent of myrtle rust (Austropuccinia psidii (synonym Puccinia psidii), Sphaerophragmiaceae, Pucciniales) was first identified as an invasive pathogen when it caused the allspice industry collapse in Jamaica in the 1930s (MacLachlan 1938), followed by the damaging impact on eucalypt industries in Brazil (Dianese et al. 1984). The first find of this disease outside its native distribution of Central and South America was made in 1977 with outbreaks on allspice in Florida (Marlatt and Kimbrough 1979). The pathogen has since spread to California, Hawaii, China, Japan, Australia, New Caledonia, South Africa, Indonesia, Singapore and New Zealand (Killgore and Heu 2007; Carnegie and Pegg 2018; EPPO 2019; Ho et al. 2019).
Austropuccinia psidii infects a range of species in the Myrtaceae family which contains over 5,500 species in 144 genera. These plants have a Gondwanan distribution and provide an important floristic component, especially in the areas where it is most species diverse (i.e. South America, Australia and Southeast Asia) (Vasconcelos et al. 2017). In the native range of myrtle rust in South America, several lineages or genetic clusters of the fungus have been found (Ross-Davis et al. 2013). Only two lineages are known to have spread out of South America: the pandemic biotype that has been found in Asia, Australia, Colombia, and the Pacific countries (du Plessis et al. 2019), and the unique biotype that to date has only been found from South Africa (Roux et al. 2016).
The host range of the A. psidii pandemic biotype has significantly expanded with its spread throughout the Pacific countries. Myrtle rust host expansion has been the most dramatic in Australia where Myrtaceae plants are diverse, widespread and abundant in native ecosystems (Myerscough 1998). Since the first detection of myrtle rust in Australia in 2010 the host range of A. psidii has rapidly increased from 129 species in 32 genera to over 480 species in 69 genera (Carnegie and Lidbetter 2012; Giblin and Carnegie 2014; Berthon et al. 2019; Fernandez Winzer et al. 2019; Soewarto et al. 2019a). While some Myrtaceae hosts are hardly affected due to resistance (Tobias et al. 2015), several plant species are severely impacted by myrtle rust and a few species in Australia face extinction (Pegg et al. 2017; Makinson 2018; Berthon et al. 2019).
In New Zealand, myrtle rust was first found in March 2017 in northernmost and isolated territory, the remote Raoul Island which is located around 1,000 km northeast of the main islands. Soon after, in May 2017, the pathogen was also found in Kerikeri, North Island (Ho et al. 2019). The pathogen was shown to be the pandemic biotype of A. psidii (du Plessis et al. 2019), the same biotype that had invaded the neighbouring countries, Australia and New Caledonia.
An incursion response was initiated immediately after the first detection by the New Zealand Ministry for Primary Industries (MPI) and continued until A. psidii was considered established to the degree that eradication was no longer feasible. In April 2018, the response was transitioned to long-term management (Ministry for Primary Industries 2018). The incursion response employed a range of resources to enable detection of the disease throughout the country and to determine which host species were affected. In this paper, we provide an overview of the diagnostic activities over the first year of response to A. psidii in New Zealand and present its distribution as well as the list of affected host species.
Material and methods
Myrtle rust notifications
A myrtle rust notification system was set up after the confirmed find of A. psidii on New Zealand mainland in May 2017. The system incorporated the notifications from members of public, government and non-government organisations, and targeted surveys by designated surveillance teams. Suspected myrtle rust finds were examined and processed by the diagnostic team at the MPI Plant Health and Environment Laboratory (PHEL).
Widespread media campaigns were released in May and September 2017 to inform the public about the disease and ask them to report suspected finds to the MPI exotic pest and disease hotline. The public were asked not to touch the plants but take images of the disease symptoms and different parts of the host plants. The calls were triaged at a call centre and the photos were pre-screened to exclude notifications that were not from Myrtaceae plants or clearly did not exhibit any rust disease symptoms. The remaining notifications were forwarded to the diagnostic team at PHEL for processing. Images received from the notifications were assessed by scientists to identify the host species, determine if the plants were infected with myrtle rust, and decide whether a sample was required for further laboratory testing. Inconclusive cases were forwarded to the survey teams to visit the site and gather more images and samples as needed.
The response surveyors were trained to recognise Myrtaceae species and myrtle rust disease symptoms, take informative images, collect samples for laboratory testing, and follow rigorous containment and decontamination procedures. They examined Myrtaceae plants in nurseries, designated high risk areas, conservation land, home gardens and public areas as required. Images of all suspected myrtle rust finds by survey teams were sent directly to the PHEL diagnostic team for processing.
Sample collection and diagnostic processing
In the beginning of the myrtle rust response, all suspected infected plant material was collected and sent to PHEL to confirm the presence of A. psidii. Symptomatic material was collected for disease confirmation and healthy plant material was collected for host identification. Samples were placed in double sealed bags, which were disinfected and securely packaged for sending to the laboratory. Upon arrival, the sample boxes were opened in a secure containment facility, symptomatic and healthy plant tissue was sampled for DNA extraction, and the DNA was stored at −80 °C. Uredinia from symptomatic plants were also used for preparing slides for morphology based identification of A. psidii. The remaining plant material was autoclaved and disposed of once the results were finalised and the sample was no longer needed.
After the identity of A. psidii was confirmed with laboratory testing on a range of host plants and the disease had been found in a number of geographical regions of New Zealand, samples were collected and tested only if they were found in new areas, on suspected new hosts or if images of symptoms did not allow conclusive identification. Apart from the above circumstances, myrtle rust and its host plant were identified based on images by the trained staff in the diagnostic team. While there are other rust fungi that infect Myrtaceae species (Simpson et al. 2006), only A. psidii is known to be present in New Zealand to date and therefore the image based identification was considered to be a reliable method.
To reduce the risk of the spread of spores and to simplify the sample processing at PHEL, the initial leaf sampling step for DNA extraction was performed in-field. Survey teams sampled four 5 × 5 mm sections from both infected and healthy leaves and separately placed these in 2 ml sample tubes pre-filled with DNA extraction buffer and metallic beads. The tubes were thereafter disinfected, placed in multiple layers of sealed bags, and further disinfected before secure packaging for sending to the laboratory.
Herbarium samples were pressed and dried to preserve the A. psidii infection on a selection of host plants whenever possible. These samples were deposited at the New Zealand Fungarium (PDD) under the voucher numbers specified in Suppl. Table 1.
DNA extraction and PCR based identification of A. psidii
The samples were macerated in 600 μl of cetyltrimethylammonium bromide (CTAB) lysis buffer (2.0% CTAB, 1.4 M NaCl, 20 mM EDTA, 100 mM Tris-HCl [pH 8.0], and 1.0% polyvinylpyrolidone) for 2 min using a Beadbeater (Biospec Products), and incubated at 65 °C for 25 min, with occasional shaking for 20 s at 2 min intervals using a Thermomixer (Eppendorf). Subsequently, the samples were centrifuged at 14,500 g for 3 min, and 420 μl of supernatant was used to perform DNA extraction using the automated KingFisher (Thermo Scientific) nucleic acid extraction system with the InviMag plant DNA mini kit (Invitek), according to the manufacturer’s instructions.
Infection with A. psidii was confirmed using a TaqMan real-time PCR assay developed by Baskarathevan et al. (2016). The qPCR assay was carried out in a final volume of 20 μl: 10 μl of 2× ToughMix (Quanta Biosciences), a final concentration of 300 nM each PpsiITS1F and PpsiITS1R primer, 120 nM FAM-labelled probe PpsiITS1P, MgCl2 (4.2 mM final), and 2 μl of DNA template. The assay was multiplexed with plant internal control, adding 150 nM each COX-F and COX-R primers, and 100 nM probe COX-P; Weller et al. 2000), Thermocycling conditions were as follows: 95 °C for 3 min, followed by 40 cycles of 95 °C for 10 s and 59 °C for 30 s.
Sequence based identification of host plants
Three regions of the host genome were amplified from the extracted DNA, the internal transcribed spacer (ITS) (primers ITS1/ITS4, White et al. 1990), external transcribed spacer (ETS) (primers Myrt (F)/ETS–18S (R), Lucas et al. 2007 and Wright et al. 2001, respectively), and the matK gene (K–J Kim unpublished primers, described in Jeanson et al. 2011). The ITS region (700 bp) was amplified using Protocol 1 as described by Buys et al. (2016) by adding 3 μl of DNA template to PCR reactions containing 1× JumpStart™ REDTaq® ReadyMix, 10 μg bovine serum albumin and primers to a final concentration of 0.3 μM. The ETS and matK regions (500 bp and 900 bp, respectively) were amplified using Buys et al. (2016) Protocol 2 by adding 2 μl of DNA template to PCR reactions containing 4 μl of HOT FIREPol® Blend Master Mix Ready to Load and primers to a final concentration of 0.3 μM. Thermocycling conditions were as follows: 94 °C for 3 min, followed by 35 cycles of 94 °C for 30 s, 56 °C for 30 s, 72 °C for 1 min and a final 10 min extension step at 72 °C.
PCR products were treated with Exonuclease I and FastAP Thermosensitive Alkaline Phosphatase (Fermentas) prior to sequencing, following the manufacturer’s instructions. DNA sequencing of the obtained amplicons was performed in both directions with forward and reverse primers on a 3500 Genetic Analyzer machine (Applied Biosystems®) using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems®). Trimmed sequences were compared against barcode reference databases of ITS, ETS and matK sequences from over 100 Myrtaceae species found in New Zealand (Buys et al. 2016) using the BLAST search tool in Geneious 10.2.5 (http://www.geneious.com). Samples that contained gene sequences from multiple species (e.g. one gene region showed 99–100% identity to one species but another gene region showed 99–100% identity to a different species) were denoted as hybrids. In all cases where this was detected, natural hybrids of the ‘parent’ species are known to occur and are common within the nursery trade.
Plant material tracing
As A. psidii can be accidentally spread through the movement of infected or contaminated material (e.g. nursery stock, plant cuttings, flowers, equipment, clothing), potential pathways both onto and off the initial known infected sites, as well as sites potentially infected by association, were intensively traced for backward and forward movement. The questionnaire used captured information on the origin and distribution of hosts grown on the property, incidence of disease and disposal methods, movement of plant material, people, vehicles and machinery, weather events, nursery management (fungicide and insecticide applications) and hygiene practices. The data collected through the questionnaires was analysed to determine if there were any links between the first detected infection site and following finds that would suggest human mediated spread.
The possibility that the pathogen was introduced to New Zealand by means other than wind dispersal was also investigated. For that, the following aspects of previous importations of Myrtaceae seeds into New Zealand were analysed: determining the proximity of the source of the seed to known myrtle rust infested areas at the time of harvest; determining their entry pathway to New Zealand (passenger, mail or cargo); and identifying where in New Zealand and by whom this material was germinated.
Results
Myrtle rust notifications
Nearly 7,000 calls reporting myrtle rust were processed by the MPI exotic pest and disease hotline during the first year of myrtle rust response in New Zealand. The peaks in the number of calls received at the call centre correlated with the media releases in May and September 2017. Nearly 40% of the calls did not report myrtle rust and were excluded either because they reported other ruts fungi on non-Myrtaceae hosts or the symptoms were not indicative of rust disease. From the remaining 60% of the notifications that were passed on to the diagnostic team at PHEL, around 30% (over 1,200 cases) were confirmed as positive for myrtle rust (Fig. 1). While at the beginning of the response, most of the calls from the public were not reporting myrtle rust, nearly half of the callers reported positive finds by April 2018. The percentage of positive finds by the trained survey team was significantly higher when compared to the notifications from members of the public and a similar increase in the efficiencies was found over time. During the first 7 months, about 40% of survey finds were confirmed positive whereas nearly 90% of the survey notifications were confirmed to represent myrtle rust during the last 6 months of the survey.
While only some of the calls from members of the public were reporting myrtle rust, the cases where they had found myrtle rust were crucial. The first notifications from the first seven known infected areas in New Zealand were all made by the public and the new areas found after that were detected by targeted surveys based on previous rust distribution modelling results. Twenty-three of the 30 New Zealand myrtle rust hosts were first found by survey teams and the remaining seven were first reported by the public. Of the latter, three were found by nurseries and four by private property owners.
Seasonal dynamics in disease expression were observed with very few new finds during the winter period (June to October) and significantly more detections during the summer months (November to May). While the increased number of positive detections in February and March was partly due to increased survey activities in infected areas, a significant increase in positive detections reported from the public was also recorded from March to May, 2018 (Fig. 1).
Pathogen and host diagnostics
In the first 5 months of the myrtle rust response, the diagnostics of myrtle rust and identification of host plants were confirmed based on examination of disease symptoms and signs and morphological characters, respectively, and molecular testing as described above. Thereafter, identification of myrtle rust and its host plants was based on images taken in the field, except for new host or new regional distribution records, or when identification was inconclusive using the images. The majority of the rust infections were confirmed based on images as the symptoms were characteristic. The greatest challenge with image based rust and host diagnostics was to obtain high quality images of symptoms and plant parts that would be characteristic for identification. New records and inconclusive cases were followed up with real-time PCR testing for A. psidii and DNA barcode analysis for the identification of host plants.
A range of visual disease symptoms were found on different host plants (Fig. 2). While some of the hosts exhibited chlorotic area around the infection sites and rust sori (e.g. Lophomyrtus bullata, Metrosideros robusta, Syzygium australe), lesions were not seen on others (e.g. Acca sellowiana). In most cases, infections were found on leaves, but on some of the hosts rust sori were also found on juvenile stems and shoot tips (e.g. L. bullata, Lophomyrtus obcordata, Metrosideros perforata, S. australe), flowers and fruits (e.g. Myrtus communis, Syzygium jambos, Ugni molinae). While the uredinial stage was dominant during the surveys, a few telia of A. psidii were found on exotic host species.
Symptoms on new and most common hosts of Austropuccinia psidii in New Zealand. New host records are marked with an asterisk. aAcca sellowiana; bEucalyptus globoidea; cLophomyrtus bullata; dLophomyrtus obcordata; eL. bullata/ L. obcordata hybrid; fMelaleuca salignus; gMelaleuca viminalis; hMetrosideros bartlettii*; iMetrosideros diffusa*; jMetrosideros excelsa; kMetrosideros fulgens*; lMetrosideros perforata*; mMetrosideros robusta*; nSyzygium australe; oSyzygium maire*
Austropuccinia psidii distribution in New Zealand
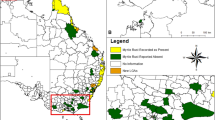
Shortly after the first detection of myrtle rust from Kerikeri, Northland on 3 May 2017, infections were also found from Taranaki (17 May) and Waikato (22 May) regions and from the Bay of Plenty area (11 June). Only a few new detections were made during the following winter months and there were no new area finds until November when infected plants were found in Auckland and Wellington; these were first detections for these regions. Infections from five additional regions were detected in March and April 2018, but the number of infected plants in these areas remained low and only a few affected hosts were found despite detailed surveys in most of these areas. The Taranaki and Bay of Plenty regions had the most myrtle rust finds throughout the response period, followed by Auckland and Waikato regions (Fig. 3).
Myrtle rust detections in New Zealand from May 2017 to June 2018. The geographic regions of New Zealand (as per Crosby et al. 1998) where Austropuccinia psidii was detected in this time period are highlighted in green. Numbers in the coloured circles indicate the total number of infected plants found in each affected region during the three different time periods (May to September 2017; October 2017 to February 2018; and March to June 2018). The date underneath the coloured circles indicates the first detection of A. psidii in the region
Host range
To date, A. psidii has been confirmed on 30 host plants in New Zealand (Table 1): 17 native taxa and 13 exotic taxa. The most frequently encountered hosts were the native L. bullata and its hybrids with L. obcordata, the native species M. excelsa, and the exotic host S. australe. This study adds 11 new taxa to the global host list: ten native taxa, most of which belonged to Metrosideros and included four hybrids; and one exotic host, a Syzygium australe x S. paniculatum hybrid. The first report of naturally occurring infections on three species were also recorded, these were Eucalyptus globoidea, Leptospermum scorparium and Thryptomene calycina. No detection of A. psidii were found on any native species of Kunzea or Neomyrtus, two other native Myrtaceae genera in New Zealand.
Discussion
Following the arrival of myrtle rust in Eastern Australia in 2010, New Zealand established a readiness plan for the potential arrival of A. psidii (Quinn and Buys 2014). The lack of an adequately resourced and coordinated response involving all key stakeholders in the early stages of the response has been criticised as one of the major gaps in the national myrtle rust response in Australia (Carnegie and Pegg 2018). With this in mind, the New Zealand response to A. psidii sought to include the general public, Māori, community groups, non-government organisations, local, regional and central government, industries, businesses and the science community where possible to ensure the best possible outcome in tracking the pathogen and employing all interested parties for a common cause. Myrtle rust survey outcomes clearly demonstrated the value of including the general public in biosecurity surveillance. Media releases during the response encouraged people to report rust like symptoms and were correlated with public reporting. While a large number of calls from public reported symptoms that were not caused by rust fungi or were on non-Myrtaceae hosts, a number of callers made a significant contribution to surveillance by either reporting new distribution areas or new hosts.
An extensive surveillance programme was deployed at the start of the myrtle rust incursion response in New Zealand which was able to effectively track the spread of the pathogen in different parts of the country and on a variety of host plants. The survey teams, trained for host and symptom recognition, mainly targeted known infected areas where they delimited the zone of infection, but also areas that were predicted to be at high infection risk in modelling studies (Beresford et al. 2018). Limited surveillance were conducted in low risk areas. The survey teams made substantial contributions by finding new host associations and a few new infected areas.
During the first year, myrtle rust spread throughout most of the North Island and upper areas of the South Island, across the areas that were predicted to be at high risk in modelling studies (Beresford et al. 2018). No evidence of potential human assisted introduction to the country (such as imported plant material) was found, which further supports the hypothesis that myrtle rust was introduced to New Zealand via air movement from Australia (Beresford et al. 2018). The detection of A. psidii in New Zealand coincided with the seasonal mass distribution of myrtaceous plants from commercial nurseries for contracted planting programmes. Movement of plant material between commercial nurseries the length of New Zealand (both North and South Islands) was common, with many transfers having already occurred prior to the first detection of A. psidii in Kerikeri, when movement controls were established. Plant movement has been estimated to be a major factor in spreading A. psidii in Australia (Carnegie and Cooper 2011; Pegg et al. 2013) and some plant material related spread may have also been possible within New Zealand. However, due to strict movement controls that were established after the detection of the fungus, most of the A. psidii spores in New Zealand were likely disseminated via air currents. Once myrtle rust spores were present in commercial and natural environments of New Zealand, the spread of the pathogen by human movement and naturally assisted distribution was inevitable, such as has occurred elsewhere (e.g. Carnegie and Cooper 2011; Pegg et al. 2013; Roux et al. 2016).
The in-field sample collection method that was introduced at the outset of the response, worked well. The risk of unintentional spread of airborne spores both in-field and in the laboratory was largely reduced when samples were collected in sealed tubes in the field, ready for DNA extraction in the laboratory. The time required to process the sample in the laboratory was also reduced. Once image based identification was determined to be sufficient for infection confirmation, the need to process samples in the laboratory was further reduced. Therefore, such a method could be recommended for handling plant material in future responses to air borne plant pathogens.
This paper presents the first complete report of the A. psidii host range in New Zealand. The high number of native species that are susceptible is concerning, as is the presence and infection of several exotic Myrtaceae species that are known to be highly susceptible overseas, such as species of Syzygium (Makinson 2018; Berthon et al. 2019). Although the number of plants infected from the survey data can give some indication of susceptibility, caution is required. Species belonging to genera such as Kunzea and Neomyrtus cannot be considered resistant based on an absence of detection. Infection and expression of disease symptoms requires the combination of pathogen, susceptible host and conducive environment, and the absence of detection may be due to the lack of inoculum exposure or suitable environmental conditions. Resistance screening on Kunzea robusta has shown that this species is susceptible to A. psidii although the population tested did have more resistance than some of the other New Zealand Myrtaceae species in which genetic resistance was either low or non-existent (Smith et al. 2020). Lophomyrtus bullata and hybrids with L. obcordata were the most frequently detected infected plant species and both resistance screening and field surveillance have indicated these to be highly susceptible (Smith et al. 2020; Soewarto et al. 2019b).
The discovery of A. psidii on A. sellowiana in New Zealand was surprising as this species was considered resistant to the pandemic biotype based on the lack of natural infection in countries where the host is grown, and the inability of the pathogen to infect A. sellowiana when artificially inoculated (Morin et al. 2012; Soewarto et al. 2019a). To date, the only reported infection of A. sellowiana by A. psidii under natural conditions was from Brazil (Blum and Dianese 2001), however, as the pandemic biotype is not known to be present in Brazil, this infection is believed to be caused by other A. psidii strains. The other detection of note was the first confirmed natural infection of L. scoparium, a host of particular concern considering the dependence of New Zealand’s mānuka honey industry on this species. Despite a number of plants being surveyed until May 2018, only one plant was found to be infected in nature. Artifical inoculation studies have, however, shown that the species is susceptible (Smith et al. 2020) which suggests that physiological or environmental factors in the field may be limiting the myrtle rust infections in mānuka plantations, as reported for other Myrtaceae hosts in Australia (Makinson 2018).
Myrtle rust has an unusually large host range for a rust fungus. Most rust fungi are highly host specific and very few have a wide host range, infecting many species within a family (Eshed and Dinoor 1981; Rytter et al. 1984; Skinner and Stuteville 1995). The ability of A. psidii to infect a wide range of species in the Myrtaceae family is therefore unique and different biotypes differ in their virulence towards different host plants (Granados et al. 2017). The already wide host range of myrtle rust and its impact to already known hosts could further expand if additional strains were introduced to the areas where the pandemic strain is currently found. For example, the dramatically increased host range in Florida over 30 years was suggested to have occurred due to importation of additional strains (Loope and La Rosa 2008) and the biotype found in South Africa has already been found to be pathogenic towards some of New Zealand’s native Myrtaceae (Soewarto et al. 2019a). To prevent introduction of additional strains to New Zealand, A. psidii remains as a quarantine organism despite establishment of the pandemic biotype. A global effort is required to prevent the spread of additional strains of A. psidii worldwide and is critical for the preservation of Myrtaceae, particularly in the Asia-Pacific region.
While the spread of myrtle rust in New Zealand was rapid and the active response moved into long-term management in April 2018, A. psidii is still very much a focus for Māori, central and local government, community groups, Myrtaceae based industries, and research institutions. Several research programmes that are currently in progress are looking for management and resistance breeding options to protect local Myrtaceae diversity and minimize the potential damage from myrtle rust to New Zealand’s unique ecosystems and industries reliant on Myrtaceae.
Change history
28 March 2020
The original version of this article unfortunately contained a mistake. The image order in Fig. 2 was incorrect. The corrected figure is given below.
References
Baskarathevan J, Taylor RK, Ho W, McDougal RL, Shivas RG, Alexander BJR (2016) Real-time PCR assay for the detection of Puccinia psidii. Plant Dis 100:617–624. https://doi.org/10.1094/PDIS-08-15-0851-RE
Beresford RM, Turner R, Tait A, Paul V, Macara G, Yu ZD, Lima L, Martin R (2018) Predicting the climatic risk of myrtle rust during its first year in New Zealand. N Z Plant Protect 71:332–347. https://doi.org/10.30843/nzpp.2018.71.176
Berthon KA, Winzer LF, Sandhu K, Cuddy W, Manea A, Carnegie AJ, Leishman MR (2019) Endangered species face an extra threat: susceptibility to the invasive pathogen Austropuccinia psidii (myrtle rust) in Australia. Australas Plant Path 48(4):385–393. https://doi.org/10.1007/s13313-019-00640-4
Blum LEB, Dianese JC (2001) Padrões de liberação de urediniósporos e desenvolvimento da ferrugem do jambeiro. Pesq Agrop Brasileira 36(6):845–850
Buys MH, Flint HJ, Miller EM, Yao H, Caird AR, Ganley RJ (2016) Preparing for the invasion: efficacy of DNA barcoding to discern the host range of myrtle rust (Puccinia psidii) among species of Myrtaceae. For Int J For Res 89(3):263–270. https://doi.org/10.1093/forestry/cpw017
Carnegie AJ, Cooper K (2011) Emergency response to the incursion of an exotic myrtaceous rust in Australia. Australas Plant Pathol 40:346. https://doi.org/10.1007/s13313-011-0066-6
Carnegie AJ, Lidbetter JR (2012) Rapidly expanding host range of Puccinia psidii sensu lato in Australia. Australas Plant Path 41:13–29. https://doi.org/10.1007/s13313-011-0082-6
Carnegie AJ, Pegg GS (2018) Lessons from the incursion of myrtle rust in Australia. Annu Rev Phytopathol 56:457–478. https://doi.org/10.1146/annurev-phyto-080516-035256
Crosby TK, Dugdale JS, Watt JC (1998) Area codes for recording specimen localities in the New Zealand subregion. N Z J Zool 25:175–183. https://doi.org/10.1080/03014223.1998.9518148
Dianese JC, Morales TSA, Silva AR (1984) Response of Eucalyptus species to field infection by Puccinia psidii. Plant Dis 68(4):314–316. https://doi.org/10.1094/PD-68-314
du Plessis E, Granados GM, Barnes I, Ho WH, Alexander BRJ, Roux J, McTaggart AR (2019) The pandemic strain of Austropuccinia psidii causes myrtle rust in New Zealand and Singapore. Australas Plant Pathol 48(3):253–256. https://doi.org/10.1007/s13313-019-0624-x
EPPO (2019) EPPO Global Database – Austropuccinia psidii. https://gd.eppo.int/taxon/PUCCPS/categorization. Accessed 1 November 2019
Eshed N, Dinoor A (1981) Genetics of pathogenicity in Puccinia coronata: the host range among grasses. Phytopathol 71:156–163
Fernandez Winzer L, Berthon KA, Carnegie AJ, Pegg GS, Leishman MR (2019) Austropuccinia psidii on the move: survey based insights to its geographical distribution, host species, impacts and management in Australia. Biol Invasions 21(4):1215–1225. https://doi.org/10.1007/s10530-018-1891-0
Giblin F & Carnegie AJ (2014) Puccinia psidii (Myrtle Rust) – Global host list. Version current on 24 Sept. 2014. http://www.anpc.asn.au/resources/Myrtle_Rust.html
Granados GM, McTaggart AR, Barnes I, Rodas CA, Roux J, Wingfield MJ (2017) The pandemic biotype of Austropuccinia psidii discovered in South America. Australas Plant Pathol 46:267–275. https://doi.org/10.1007/s13313-017-0488-x
Ho WWH, Baskarathevan J, Griffin RL, Quinn B, Alexander BJR, Havell D, Ward N, Pathan A (2019) First report of myrtle rust caused by Austropuccinia psidii on Metrosideros kermadecensis on Raoul Island and on M. excelsa in Kerikeri, New Zealand. Plant Dis 103(8):2128. https://doi.org/10.1094/PDIS-12-18-2243-PDN
Jeanson ML, Labat JN, Little DP (2011) DNA barcoding: a new tool for palm taxonomists? Ann Bot 108(8):1445–1451. https://doi.org/10.1093/aob/mcr158
Killgore EM, Heu RA (2007) Ohia rust Puccinia psidii Winter. State of Hawaii Department of Agriculture, New Pest Advisory No 05–04. https://hdoa.hawaii.gov/pi/files/2013/01/npa05-04-ohiarust.pdf
Loope L, La Rosa AM (2008) An analysis of the risk of introduction of additional strains of the rust Puccinia psidii Winter (‘ōhi‘a rust) to Hawai‘i. U.S. Geological Survey Open File Report 2008–1008. 11 p. http://pubs.usgs.gov/of/2008/1008/. Accessed 4 November 2019
Lucas EJ, Harris S, Mazine F, Belsham S, Lughadha E, Telford A, Gasson P, Chase M (2007) Suprageneric phylogenetics of Myrteae, the generically richest tribe in Myrtaceae (Myrtales). Taxon 56:1105–1128. https://doi.org/10.2307/25065906
MacLachlan JD (1938) A rust of the pimento tree in Jamaica, BWI. Phytopathology 28:157–170
Makinson RO (2018) Myrtle rust reviewed: the impacts of the invasive plant pathogen Austropuccinia psidii on the Australian environment. Plant Biosecurity Cooperative Research Centre, Canberra. http://www.apbsf.org.au/wp-content/uploads/2018/11/Myrtle-Rust-reviewed-June-22-2018-web.pdf
Marlatt RB, Kimbrough JW (1979) Puccinia psidii on Pimenta dioica in south Florida. Plant Dis Rep 63(6):510–512
Ministry for Primary Industries (2018) New approach to manage myrtle rust. MPI media release published on 06 April 2018. https://www.mpi.govt.nz/news-and-resources/media-releases/new-approach-to-manage-myrtle-rust/
Morin L, Aveyard R, Lidbetter JR, Wilson PG (2012) Investigating the host-range of the rust fungus Puccinia psidii sensu lato across tribes of the family Myrtaceae present in Australia. PLoS One 7(4):e35434. https://doi.org/10.1371/journal.pone.0035434
Myerscough PJ (1998) Ecology of Myrtaceae with special reference to the Sydney region. Cunninghamia 5(4):787–807
Pegg GS, Giblin FR, McTaggart AR, Guymer GP, Taylor H, Ireland KB, Shivas RG, Perry S (2013) Puccinia psidii in Queensland, Australia: disease symptoms, distribution and impact. Plant Pathol 63(5):1105–1021. https://doi.org/10.1111/ppa.12173
Pegg G, Taylor T, Entwistle P, Guymer G, Giblin F, Carnegie A (2017) Impact of Austropuccinia psidii (myrtle rust) on Myrtaceae rich wet sclerophyll forests in south East Queensland. PLoS One 12(11):e0188058. https://doi.org/10.1371/journal.pone.0188058
Quinn B, Buys MH (2014) Getting ready for myrtle rust. Surveillance 41(4):27–28 https://www.mpi.govt.nz/dmsdocument/5002-surveillance-magazine-vol-41-no-4-december-2014
Ross-Davis AL, Graça RN, Alfenas AC, Peever TL, Hanna JW, Uchida JY, Hauff RD, Kadooka CY, Kim MS, Cannon PG, Namba S, Minato N, Simeto S, Pérez CA, Rayamajhi MB, Móran M, Lodge DJ, Arguedas M, Medel-Ortiz R, López-Ramirez A, Tennant P, Glen M, Klopfenstein NB (2013) Tracking the distribution of Puccinia psidii genotypes that cause rust disease on diverse Myrtaceous trees and shrubs. In: http://www.Fs.Fed.Us/rm/pubs_journals/2014/rmrs_2014_ross_davis_a001.Pdf. Accessed 1 November 2019
Roux J, Granados GM, Shuey L, Barnes I, Wingfield MJ, McTaggart AR (2016) A unique genotype of the rust pathogen, Puccinia psidii, on Myrtaceae in South Africa. Australas Plant Path 45:645–652. https://doi.org/10.1007/s13313-016-0447-y
Rytter JL, Dowler WM, Bromfield KR (1984) Additional alternative hosts of Phakopsora pachyrhizi, causal agent of soybean rust. Plant Dis 68:818–819
Simpson JA, Thomas K, Grgurinovic CA (2006) Uredinales species pathogenic on species of Myrtaceae. Australas Plant Pathol 35:549–562. https://doi.org/10.1071/AP06057
Skinner DZ, Stuteville DL (1995) Host range expansion of the alfalfa rust pathogen. Plant Dis 79:456–460
Smith RG, Ganley BJ, Chagné D, Nadarajan J, Pathirana RN, Ryan J, Arnst EA, Sutherland R, Soewarto J, Houliston G, Marsh AT, Koot E, Carnegie A, Menzies T, Lee DJ, Shuey LS, Pegg GS (2020) Resistance of New Zealand provenance Leptospermum scoparium, Kunzea robusta, Kunzea linearis and Metrosideros excelsa to Austropuccinia psidii. Plant Dis. https://doi.org/10.1094/PDIS-11-19-2302-RE
Soewarto J, Giblin F, Carnegie AJ (2019a) Austropuccinia psidii (myrtle rust) global host list. Version 2. Australian network for plant conservation, Canberra, ACT. http://www.anpc.asn.au/myrtle-rust. Accessed 4 November 2019
Soewarto J, Sutherland R, Ganley B, du Plessis E, Barnes I, Wingfield M, Granados G (2019b). Assessment of other myrtle rust biotypes. Ministry for Primary Industries 18608 project report. https://www.myrtlerust.org.nz/assets/Uploads/Assessment-of-other-myrtle-rust-biotypes.pdf. Accessed 4 November 2019
Tobias PA, Guest DI, Külheim C, Hsieh J-F, Park RF (2015) A curious case of resistance to a new encounter pathogen: myrtle rust in Australia. Mol Plant Pathol 17(5):783–788. https://doi.org/10.1111/mpp.12331
Vasconcelos TNC, Proença CEB, Ahmad B, Aguilar DS, Aguilar R, Amorim BS, Campbell K, Costa IR, De-Carvalho PS, Faria JEQ, Giaretta A, Kooij PW, Lima DF, Mazine FF, Peguero B, Prenner G, Santos MF, Soewarto J, Wingler A, Lucas EJ (2017) Myrteae phylogeny, calibration, biogeography and diversification patterns: increased understanding in the most species rich tribe of Myrtaceae. Mol Phylogenet Evol 109:113–137. https://doi.org/10.1016/j.ympev.2017.01.002
Weller SA, Elphinstone JG, Smith NC, Boonham N, Stead DE (2000) Detection of Ralstonia solanacearum strains with a quantitative, multiplex, real-time, fluorogenic PCR (TaqMan) assay. Appl Environ Microbiol 66(7):2853–2858. https://doi.org/10.1128/aem.66.7.2853-2858.2000
White T, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal DNA genes for phylogenetics. In: Innis M, Gelfand D, Snrnsky J, White T (eds) PCR Protocols. Academic Press, New York, pp 315–322
Wright SD, Yong CG, Wichman SR, Dawson JW, Gardner RC (2001) Stepping stones to Hawaii: a trans-equatorial dispersal pathway for Metrosideros (Myrtaceae) inferred from nrDNA (ITS+ ETS). J Biogeogr 28(6):769–774. https://doi.org/10.1046/j.1365-2699.2001.00605.x
Acknowledgements
We thank: Wendy Blount, Katharina Hofer, Santha France, Stephanie Keeling, Stacey Lamont, Michael Surrey and the AsureQuality staff for managing the notifications from the public; Ruth Griffin, Jeyaseelan Baskarathevan, Subuhi Khan, Julie Pattemore, Zoila Perez and Rob Taylor for carrying out the A. psidii PCR testing; Diane Anderson, Jeyaseelan Baskarathevan, Disna Gunawardana, Benedicte Lebas and Rob Taylor who also contributed to leading various diagnostic aspects of the response; Jane Martin for handling logistics in the laboratory; Phill Sherring for providing the MPI media release data; Mei Wang for sharing the data from the MPI Exotic Pest and Disease hotline call centre; surveillance teams for collecting the samples and taking images; Scion staff who contributed to host identification; and Emma Scheltema for figure design. Rob Taylor, Lia Liefting, two anonymous reviewers and the editor provided valuable suggestions to improve the previous versions of this manuscript.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(DOCX 15 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Toome-Heller, M., Ho, W.W.H., Ganley, R.J. et al. Chasing myrtle rust in New Zealand: host range and distribution over the first year after invasion. Australasian Plant Pathol. 49, 221–230 (2020). https://doi.org/10.1007/s13313-020-00694-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-020-00694-9