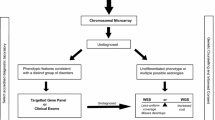
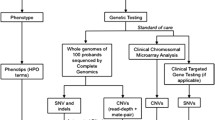
Abstract
Chromosomal microarray and Next-generation sequencing are two widely used genomic tests that have improved the diagnosis of children with a genetic condition. Chromosomal microarray has become a first-tier test in evaluating children with intellectual disability, multiple malformations and autism due to its higher yield and resolution. Next generation sequencing, that includes targeted panel testing, exome sequencing and whole genome sequencing ends diagnostic odyssey in 25–30% of unselected children with rare monogenic syndromes, especially when the condition is genetically heterogeneous. This article provides a review of these genomic tests for pediatricians.
Similar content being viewed by others
References
Bernardini L, Alesi V, Loddo S, Novelli A, Bottillo I, Battaglia A, et al. High-resolution SNP arrays in mental retardation diagnostics: How much do we gain? Eur J Hum Genet. 2010;18:178–85.
Manning M, Hudgins L. Array-based technology and recommendations for utilization in medical genetics practice for detection of chromosomal abnormalities. Genet Med. 2010;12: 742–5.
Miller DT, Adam MP, Aradhya S, Biesecker LG, Brothman AR, Carter NP, et al. Consensus Statement: Chromosomal Microarray is a First-tier Clinical diagnostic Test for Individuals With Developmental Disabilities or Congenital Aanomalies. Am J Hum Genet. 2010;86: 749–64.
Snijders AM, Nowak N, Segraves R, Blackwood S, Brown N, Conroy J, et al. Assembly of microarrays for genome-wide measurement of DNA copy number. Nat Genet. 2001;29: 263–4.
Beaudet AL. The utility of chromosomal microarray analysis in developmental and behavioral pediatrics. Child Dev. 2013;84: 121–32.
Oostlander AE, Meijer GA, Ylstra B. Microarray-based comparative genomic hybridization and its applications in human genetics. Clin Genet. 2004;66: 488–95.
LaFramboise T. Single nucleotide polymorphism arrays: A decade of biological, computational and technological advances. Nucleic Acids Res. 2009;37: 4181–93.
Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat Rev Genet. 2006;7: 85–97.
Kearney HM, Thorland EC, Brown KK, Quintero-Rivera F, South ST. American College of Medical Genetics Standards and Guidelines for Interpretation and Reporting of Postnatal Constitutional Copy Number Variants. Genet Med. 2011;13: 680–5.
South ST, Lee C, Lamb AN, Higgins AW, Kearney HM. ACMG Standards and Guidelines for Constitutional Cytogenomic Microarray Analysis, Including Postnatal and Prenatal applications: Revision 2013. Genet Med. 2013;15: 901–9.
Levy B, Wapner R. Prenatal diagnosis by chromosomal microarray analysis. Fertil Steril. 2018;109: 201–12.
Rauch A, Hoyer J, Guth S, Zweier C, Kraus C, Becker C, et al. Diagnostic yield of various genetic approaches in patients with unexplained developmental delay or mental retardation. Am J Med Genet A. 2006;140: 2063–74.
Karampetsou E, Morrogh D, Chitty L. Microarray technology for the diagnosis of fetal chromosomal aberrations: Which platform should we use? J Clin Med. 2014;3:663–78.
Edelmann L, Hirschhorn K. Clinical utility of array CGH for the detection of chromosomal imbalances associated with mental retardation and multiple congenital anomalies. Ann NY Acad Sci. 2009;1151: 157–66.
Sismani C, Kitsiou-Tzeli S, Ioannides M, Christodoulou C, Anastasiadou V, Stylianidou G, et al. Cryptic genomic imbalances in patients with de novo or familial apparently balanced translocations and abnormal phenotype. Mol Cytogenet. 2008;1: 15.
Thiffault I, Lantos J. The challenge of analyzing the results of next-generation sequencing in children. Pediatrics. 2016;137: S3–7.
Richards S, Aziz N, Bale S, Bick D, Das S, Gastier-Foster J, et al. Standards and Guidelines for the Interpretation of Sequence Variants: A Joint Consensus Recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet Med. 2015;17: 405–24.
Biesecker LG, Biesecker BB. An approach to pediatric exome and genome sequencing. Curr Opin Pediatr. 2014;26: 639–45.
Biesecker LG, Green RC. Diagnostic clinical genome and exome sequencing. N Engl J Med. 2014;371: 1170.
Schwarze K, Buchanan J, Taylor JC, Wordsworth S. Are whole-exome and whole-genome sequencing approaches cost-effective? A systematic review of the literature. Genet Med. 2018;20: 1122–30.
Yang Y, Muzny DM, Reid JG, Bainbridge MN, Willis A, Ward PA, et al. Clinical whole-exome sequencing for the diagnosis of mendelian disorders. N Engl J Med. 2013;369: 1502–11.
Author information
Authors and Affiliations
Contributions
DLN: substantial contributions to design and draft of the work; GKM: substantial contributions to the conception and design of the work, drafting and revising it critically for important intellectual content. Both approve the final version to be published and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Corresponding author
Rights and permissions
About this article
Cite this article
Narayanan, D.L., Girisha, K.M. Genomic Testing for Diagnosis of Genetic Disorders in Children: Chromosomal Microarray and Next—Generation Sequencing. Indian Pediatr 57, 549–554 (2020). https://doi.org/10.1007/s13312-020-1853-3
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13312-020-1853-3