Abstract
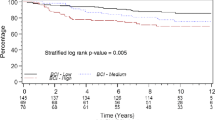
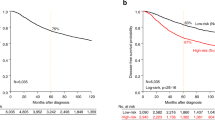
Breast carcinoma (BC) corresponds to 23 % of all cancers in women, with 1.38 million new cases and 460,000 deaths worldwide annually. Despite the significant advances in the identification of molecular markers and different modalities of treatment for primary BC, the ability to predict its metastatic behavior is still limited. The purpose of this study was to identify novel molecular markers associated with distinct clinical outcomes in a Brazilian cohort of BC patients. We generated global gene expression profiles using tumor samples from 24 patients with invasive ductal BC who were followed for at least 5 years, including a group of 15 patients with favorable outcomes and another with nine patients who developed metastasis. We identified a set of 58 differentially expressed genes (p ≤ 0.01) between the two groups. The prognostic value of this metastasis signature was corroborated by its ability to stratify independent BC patient datasets according to disease-free survival and overall survival. The upregulation of B3GNT7, PPM1D, TNKS2, PHB, and GTSE1 in patients with poor outcomes was confirmed by quantitative reverse transcription polymerase chain reaction (RT-qPCR) in an independent sample of patients with BC (47 with good outcomes and eight that presented metastasis). The expression of BCL2-associated agonist of cell death (BAD) protein was determined in 1276 BC tissue samples by immunohistochemistry and was consistent with the reduced BAD mRNA expression levels in metastatic cases, as observed in the oligoarray data. These findings point to novel prognostic markers that can distinguish breast carcinomas with metastatic potential from those with favorable outcomes.




Similar content being viewed by others
References
Veta M, Pluim JP, van Diest PJ, Viergever MA. Breast cancer histopathology image analysis: a review. IEEE Trans Biomed Eng. 2014;61:1400–11.
Dumalaon-Canaria JA, Hutchinson AD, Prichard I, Wilson C. What causes breast cancer? A systematic review of causal attributions among breast cancer survivors and how these compare to expert-endorsed risk factors. Cancer Causes Control. 2014;25:771–85.
Colombo PE, Milanezi F, Weigelt B, Reis-Filho JS. Microarrays in the 2010s: the contribution of microarray-based gene expression profiling to breast cancer classification, prognostication and prediction. Breast Cancer Res. 2011;13:212.
Rosa M. Advances in the molecular analysis of breast cancer: pathway toward personalized medicine. Cancer Control. 2015;22:211–9.
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y, Graf S, Ha G, Haffari G, Bashashati A, Russell R, McKinney S, Langerod A, Green A, Provenzano E, Wishart G, Pinder S, Watson P, Markowetz F, Murphy L, Ellis I, Purushotham A, Borresen-Dale AL, Brenton JD, Tavare S, Caldas C, Aparicio S. The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature. 2012;486:346–52.
Reis-Filho JS, Weigelt B, Fumagalli D, Sotiriou C. Molecular profiling: moving away from tumor philately. Sci Transl Med. 2010;2:47ps43.
Taherian-Fard A, Srihari S, Ragan MA. Breast cancer classification: linking molecular mechanisms to disease prognosis. Brief Bioinform. 2015;16:461–74.
Toss A, Cristofanilli M. Molecular characterization and targeted therapeutic approaches in breast cancer. Breast Cancer Res. 2015;17:60.
Patani N, Martin LA, Dowsett M. Biomarkers for the clinical management of breast cancer: international perspective. Int J Cancer. 2013;133:1–13.
Peppercorn J, Perou CM, Carey LA. Molecular subtypes in breast cancer evaluation and management: divide and conquer. Cancer Investig. 2008;26:1–10.
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT, Schreiber GJ, Kerkhoven RM, Roberts C, Linsley PS, Bernards R, Friend SH. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–6.
Wang Y, Klijn JG, Zhang Y, Sieuwerts AM, Look MP, Yang F, Talantov D, Timmermans M, Meijer-van Gelder ME, Yu J, Jatkoe T, Berns EM, Atkins D, Foekens JA. Gene-expression profiles to predict distant metastasis of lymph-node-negative primary breast cancer. Lancet. 2005;365:671–9.
Drukker CA, Bueno-de-Mesquita JM, Retel VP, van Harten WH, van Tinteren H, Wesseling J, Roumen RM, Knauer M, van’t Veer LJ, Sonke GS, Rutgers EJ, van de Vijver MJ, Linn SC. A prospective evaluation of a breast cancer prognosis signature in the observational RASTER study. Int J Cancer. 2013;133:929–36.
Goldhirsch A, Ingle JN, Gelber RD, Coates AS, Thurlimann B, Senn HJ. Thresholds for therapies: highlights of the St Gallen International Expert Consensus on the primary therapy of early breast cancer 2009. Ann Oncol. 2009;20:1319–29.
Sotiriou C, Pusztai L. Gene-expression signatures in breast cancer. N Engl J Med. 2009;360:790–800.
Filipits M, Nielsen TO, Rudas M, Greil R, Stoger H, Jakesz R, Bago-Horvath Z, Dietze O, Regitnig P, Gruber-Rossipal C, Muller-Holzner E, Singer CF, Mlineritsch B, Dubsky P, Bauernhofer T, Hubalek M, Knauer M, Trapl H, Fesl C, Schaper C, Ferree S, Liu S, Cowens JW, Gnant M. The PAM50 risk-of-recurrence score predicts risk for late distant recurrence after endocrine therapy in postmenopausal women with endocrine-responsive early breast cancer. Clin Cancer Res. 2014;20:1298–305.
Loi S, Haibe-Kains B, Desmedt C, Lallemand F, Tutt AM, Gillet C, Ellis P, Harris A, Bergh J, Foekens JA, Klijn JG, Larsimont D, Buyse M, Bontempi G, Delorenzi M, Piccart MJ, Sotiriou C. Definition of clinically distinct molecular subtypes in estrogen receptor-positive breast carcinomas through genomic grade. J Clin Oncol Off J Am Soc Clin Oncol. 2007;25:1239–46.
Ma XJ, Salunga R, Dahiya S, Wang W, Carney E, Durbecq V, Harris A, Goss P, Sotiriou C, Erlander M, Sgroi D. A five-gene molecular grade index and HOXB13:IL17BR are complementary prognostic factors in early stage breast cancer. Clin Cancer Res. 2008;14:2601–8.
Reis-Filho JS, Pusztai L. Gene expression profiling in breast cancer: classification, prognostication, and prediction. Lancet. 2011;378:1812–23.
Ein-Dor L, Kela I, Getz G, Givol D, Domany E. Outcome signature genes in breast cancer: is there a unique set? Bioinformatics. 2005;21:171–8.
Gormley M, Dampier W, Ertel A, Karacali B, Tozeren A. Prediction potential of candidate biomarker sets identified and validated on gene expression data from multiple datasets. BMC Bioinf. 2007;8:415.
Korkola JE, Blaveri E, DeVries S, Moore 2nd DH, Hwang ES, Chen YY, Estep AL, Chew KL, Jensen RH, Waldman FM. Identification of a robust gene signature that predicts breast cancer outcome in independent data sets. BMC Cancer. 2007;7:61.
Fan C, DS O, Wessels L, Weigelt B, Nuyten DS, Nobel AB, van’t Veer LJ, Perou CM. Concordance among gene-expression-based predictors for breast cancer. N Engl J Med. 2006;355:560–9.
Haibe-Kains B, Desmedt C, Piette F, Buyse M, Cardoso F, Van’t Veer L, Piccart M, Bontempi G, Sotiriou C. Comparison of prognostic gene expression signatures for breast cancer. BMC Genomics. 2008;9:394.
Paik S. Is gene array testing to be considered routine now? Breast. 2011;20(Suppl 3):S87–91.
Saini A, Hou J, Zhou W. Breast cancer prognosis risk estimation using integrated gene expression and clinical data. Biomed Res Int. 2014;2014:459203.
Wirapati P, Sotiriou C, Kunkel S, Farmer P, Pradervand S, Haibe-Kains B, Desmedt C, Ignatiadis M, Sengstag T, Schutz F, Goldstein DR, Piccart M, Delorenzi M. Meta-analysis of gene expression profiles in breast cancer: toward a unified understanding of breast cancer subtyping and prognosis signatures. Breast Cancer Res. 2008;10:R65.
Gyorffy B, Hatzis C, Sanft T, Hofstatter E, Aktas B, Pusztai L. Multigene prognostic tests in breast cancer: past, present, future. Breast Cancer Res. 2015;17:11.
Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–93.
Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, Coller H, Loh ML, Downing JR, Caligiuri MA, Bloomfield CD, Lander ES. Molecular classification of cancer: class discovery and class prediction by gene expression monitoring. Science. 1999;286:531–7.
Hu Z, Bonifas JM, Aragon G, Kopelovich L, Liang Y, Ohta S, Israel MA, Bickers DR, Aszterbaum M, Epstein Jr EH. Evidence for lack of enhanced hedgehog target gene expression in common extracutaneous tumors. Cancer Res. 2003;63:923–8.
Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45.
Aguirre-Gamboa R, Gomez-Rueda H, Martinez-Ledesma E, Martinez-Torteya A, Chacolla-Huaringa R, Rodriguez-Barrientos A, Tamez-Pena JG, Trevino V. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS One. 2013;8:e74250.
Xie C, Mao X, Huang J, Ding Y, Wu J, Dong S, Kong L, Gao G, Li CY, Wei L. Kobas 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 2011;39:W316–22.
Nagai MA, Fregnani JH, Netto MM, Brentani MM, Soares FA. Down-regulation of phlda1 gene expression is associated with breast cancer progression. Breast Cancer Res Treat. 2007;106:49–56.
Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, d’Assignies MS, Bergh J, Lidereau R, Ellis P, Harris AL, Klijn JG, Foekens JA, Cardoso F, Piccart MJ, Buyse M, Sotiriou C. Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res. 2007;13:3207–14.
Jezequel P, Campone M, Roche H, Gouraud W, Charbonnel C, Ricolleau G, Magrangeas F, Minvielle S, Geneve J, Martin AL, Bataille R, Campion L. A 38-gene expression signature to predict metastasis risk in node-positive breast cancer after systemic adjuvant chemotherapy: a genomic substudy of PACS01 clinical trial. Breast Cancer Res Treat. 2009;116:509–20.
Karlsson E, Delle U, Danielsson A, Olsson B, Abel F, Karlsson P, Helou K. Gene expression variation to predict 10-year survival in lymph-node-negative breast cancer. BMC Cancer. 2008;8:254.
Minn AJ, Kang Y, Serganova I, Gupta GP, Giri DD, Doubrovin M, Ponomarev V, Gerald WL, Blasberg R, Massague J. Distinct organ-specific metastatic potential of individual breast cancer cells and primary tumors. J Clin Invest. 2005;115:44–55.
Molloy TJ, Roepman P, Naume B, van’t Veer LJ. A prognostic gene expression profile that predicts circulating tumor cell presence in breast cancer patients. PLoS One. 2012;7:e32426.
Sorlie T, Wang Y, Xiao C, Johnsen H, Naume B, Samaha RR, Borresen-Dale AL. Distinct molecular mechanisms underlying clinically relevant subtypes of breast cancer: gene expression analyses across three different platforms. BMC Genomics. 2006;7:127.
van de Vijver MJ, He YD, van’t Veer LJ, Dai H, Hart AA, Voskuil DW, Schreiber GJ, Peterse JL, Roberts C, Marton MJ, Parrish M, Atsma D, Witteveen A, Glas A, Delahaye L, van der Velde T, Bartelink H, Rodenhuis S, Rutgers ET, Friend SH, Bernards R. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009.
Wang DY, Done SJ, McCready DR, Boerner S, Kulkarni S, Leong WL. A new gene expression signature, the clinicomolecular triad classification, may improve prediction and prognostication of breast cancer at the time of diagnosis. Breast Cancer Res. 2011;13:R92.
Zhao H, Langerod A, Ji Y, Nowels KW, Nesland JM, Tibshirani R, Bukholm IK, Karesen R, Botstein D, Borresen-Dale AL, Jeffrey SS. Different gene expression patterns in invasive lobular and ductal carcinomas of the breast. Mol Biol Cell. 2004;15:2523–36.
TCGA. Comprehensive molecular portraits of human breast tumours. Nature. 2012;490:61–70.
Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, Baehner FL, Walker MG, Watson D, Park T, Hiller W, Fisher ER, Wickerham DL, Bryant J, Wolmark N. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. 2004;351:2817–26.
Kao KJ, Chang KM, Hsu HC, Huang AT. Correlation of microarray-based breast cancer molecular subtypes and clinical outcomes: implications for treatment optimization. BMC Cancer. 2011;11:143.
Reis-Filho JS, Westbury C, Pierga JY. The impact of expression profiling on prognostic and predictive testing in breast cancer. J Clin Pathol. 2006;59:225–31.
Li X, Jiang S, Tapping RI. Toll-like receptor signaling in cell proliferation and survival. Cytokine. 2010;49:1–9.
Luddy KA, Robertson-Tessi M, Tafreshi NK, Soliman H, Morse DL. The role of toll-like receptors in colorectal cancer progression: evidence for epithelial to leucocytic transition. Front Immunol. 2014;5:429.
Kidd LC, Rogers EN, Yeyeodu ST, Jones DZ, Kimbro KS. Contribution of toll-like receptor signaling pathways to breast tumorigenesis and treatment. Breast Cancer (Dove Med Press). 2013;5:43–51.
Powell E, Piwnica-Worms D, Piwnica-Worms H. Contribution of p53 to metastasis. Cancer Discov. 2014;4:405–14.
Kaenel P, Mosimann M, Andres AC. The multifaceted roles of Eph/ephrin signaling in breast cancer. Cell Adhes Migr. 2012;6:138–47.
Cerqueira OL, Truesdell P, Baldassarre T, Vilella-Arias SA, Watt K, Meens J, Chander H, Osorio CA, Soares FA, Reis EM, Craig AW. CIP4 promotes metastasis in triple-negative breast cancer and is associated with poor patient prognosis. Oncotarget. 2015;6:9397–408.
Pichot CS, Arvanitis C, Hartig SM, Jensen SA, Bechill J, Marzouk S, Yu J, Frost JA, Corey SJ. Cdc42-interacting protein 4 promotes breast cancer cell invasion and formation of invadopodia through activation of N-WASp. Cancer Res. 2010;70:8347–56.
Rauta J, Alarmo EL, Kauraniemi P, Karhu R, Kuukasjarvi T, Kallioniemi A. The serine-threonine protein phosphatase PPM1D is frequently activated through amplification in aggressive primary breast tumours. Breast Cancer Res Treat. 2006;95:257–63.
Demetriou M, Granovsky M, Quaggin S, Dennis JW. Negative regulation of t-cell activation and autoimmunity by Mgat5 N-glycosylation. Nature. 2001;409:733–9.
Nakamura N, Yamakawa N, Sato T, Tojo H, Tachi C, Furukawa K. Differential gene expression of beta-1,4-galactosyltransferases I, II and V during mouse brain development. J Neurochem. 2001;76:29–38.
Teneberg S, Leonardsson I, Karlsson H, Jovall PA, Angstrom J, Danielsson D, Naslund I, Ljungh A, Wadstrom T, Karlsson KA. Lactotetraosylceramide, a novel glycosphingolipid receptor for Helicobacter pylori, present in human gastric epithelium. J Biol Chem. 2002;277:19709–19.
Kataoka K, Huh NH. A novel beta1,3-N-acetylglucosaminyltransferase involved in invasion of cancer cells as assayed in vitro. Biochem Biophys Res Commun. 2002;294:843–8.
Sinclair CS, Rowley M, Naderi A, Couch FJ. The 17q23 amplicon and breast cancer. Breast Cancer Res Treat. 2003;78:313–22.
Li J, Yang Y, Peng Y, Austin RJ, van Eyndhoven WG, Nguyen KC, Gabriele T, McCurrach ME, Marks JR, Hoey T, Lowe SW, Powers S. Oncogenic properties of ppm1d located within a breast cancer amplification epicenter at 17q23. Nat Genet. 2002;31:133–4.
Lu X, Nguyen TA, Donehower LA. Reversal of the ATM/ATR-mediated DNA damage response by the oncogenic phosphatase PPM1D. Cell Cycle. 2005;4:1060–4.
Bulavin DV, Phillips C, Nannenga B, Timofeev O, Donehower LA, Anderson CW, Appella E, Fornace Jr AJ. Inactivation of the Wip1 phosphatase inhibits mammary tumorigenesis through p38 MAPK-mediated activation of the p16(Ink4a)-p19(Arf) pathway. Nat Genet. 2004;36:343–50.
Bulavin DV, Demidov ON, Saito S, Kauraniemi P, Phillips C, Amundson SA, Ambrosino C, Sauter G, Nebreda AR, Anderson CW, Kallioniemi A, Fornace Jr AJ, Appella E. Amplification of ppm1d in human tumors abrogates p53 tumor-suppressor activity. Nat Genet. 2002;31:210–5.
Demidov ON, Kek C, Shreeram S, Timofeev O, Fornace AJ, Appella E, Bulavin DV. The role of the MKK6/p38 MAPK pathway in Wip1-dependent regulation of ErbB2-driven mammary gland tumorigenesis. Oncogene. 2007;26:2502–6.
Oliva-Trastoy M, Berthonaud V, Chevalier A, Ducrot C, Marsolier-Kergoat MC, Mann C, Leteurtre F. The Wip1 phosphatase (PPM1D) antagonizes activation of the Chk2 tumour suppressor kinase. Oncogene. 2007;26:1449–58.
Monz D, Munnia A, Comtesse N, Fischer U, Steudel WI, Feiden W, Glass B, Meese EU. Novel tankyrase-related gene detected with meningioma-specific sera. Clin Cancer Res. 2001;7:113–9.
Wang S, Fusaro G, Padmanabhan J, Chellappan SP. Prohibitin co-localizes with Rb in the nucleus and recruits N-CoR and HDAC1 for transcriptional repression. Oncogene. 2002;21:8388–96.
Rajalingam K, Rudel T. Ras-Raf signaling needs prohibitin. Cell Cycle. 2005;4:1503–5.
Nijtmans LG, Artal SM, Grivell LA, Coates PJ. The mitochondrial PHB complex: roles in mitochondrial respiratory complex assembly, ageing and degenerative disease. Cell Mol Life Sci. 2002;59:143–55.
Monte M, Collavin L, Lazarevic D, Utrera R, Dragani TA, Schneider C. Cloning, chromosome mapping and functional characterization of a human homologue of murine gtse-1 (B99) gene. Gene. 2000;254:229–36.
Monte M, Benetti R, Buscemi G, Sandy P, Del Sal G, Schneider C. The cell cycle-regulated protein human gtse-1 controls DNA damage-induced apoptosis by affecting p53 function. J Biol Chem. 2003;278:30356–64.
Zhou X, Temam S, Oh M, Pungpravat N, Huang BL, Mao L, Wong DT. Global expression-based classification of lymph node metastasis and extracapsular spread of oral tongue squamous cell carcinoma. Neoplasia. 2006;8:925–32.
Cekanova M, Fernando RI, Siriwardhana N, Sukhthankar M, De la Parra C, Woraratphoka J, Malone C, Strom A, Baek SJ, Wade PA, Saxton AM, Donnell RM, Pestell RG, Dharmawardhane S, Wimalasena J. BCL-2 family protein, BAD is down-regulated in breast cancer and inhibits cell invasion. Exp Cell Res. 2015;331:1–10.
Acknowledgments
This work was mainly supported by a grant from the Conselho Nacional de Desenvolvimento Científico e Tecnologico (CNPq) (Edital MCT/CNPq/CT-Biotecnologia n° 010/2004). Additional funding was provided by the Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP). E.M.R. and S.V.A. received investigator fellowship awards from CNPq. The authors thank Sandra Dringo Linde for her expert technical assistance during this study.
Authors’ contributions
SVA, SRR, and EMR conceived and designed the experiments. RAC and MACD performed the experiments. EMR, FM, and RAC analyzed data. JRFC and VPA contributed samples. RAC, FM, VPA, SRR, and EMR drafted or revised the manuscript. All authors read and approved the final manuscript.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Written informed consent was obtained from all patients during the collection period, and the study was reviewed and approved by the Ethics Committees from both institutions (CEP FHAC 340/04 and CEP ACCC 1155/08).
Conflicts of interest
None
Additional information
Silvia R. Rogatto and Eduardo M. Reis contributed equally to this study.
Electronic supplementary material
Supplementary Figure 1
(PDF 445 kb)
Supplementary Figure 2
(PDF 257 kb)
Supplementary Figure 3
(PDF 50 kb)
Supplementary Figure 4
(PDF 27 kb)
Supplementary Table 1
(PDF 144 kb)
Supplementary Table 2
(PDF 149 kb)
Supplementary Table 3
(PDF 144 kb)
Supplementary Table 4
(PDF 142 kb)
Supplementary Table 5
(PDF 114 kb)
Supplementary Table 6
(PDF 258 kb)
Supplementary Table 7
(PDF 139 kb)
Rights and permissions
About this article
Cite this article
Canevari, R.A., Marchi, F.A., Domingues, M.A.C. et al. Identification of novel biomarkers associated with poor patient outcomes in invasive breast carcinoma. Tumor Biol. 37, 13855–13870 (2016). https://doi.org/10.1007/s13277-016-5133-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5133-8