Abstract
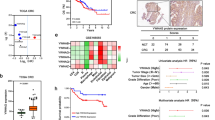
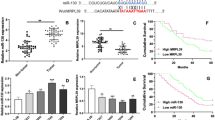
Lung cancer is the most common cause of cancer-related deaths worldwide, and non-small cell lung cancer (NSCLC) accounts for 80 to 85 % of all lung cancer. Although the standard treatment regimen has been established, long-term survival for NSCLC patients is still generally poor. The histone demethylase Jumonji domain containing 1A (JMJD1A) has been proposed as an oncogene in several types of human cancer, but its clinical significance and functional roles in NSCLC remain largely unclear. In the present study, JMJD1A was frequently upregulated in NSCLC compared with para-carcinoma tissues. JMJD1A knockdown significantly inhibited NSCLC cell growth, migration, and invasion in vitro and tumorigenesis in vivo. Further experiments demonstrated that JMJD1A knockdown could decrease the expression of EZH2, which has been shown to play a crucial role in the carcinogenesis of NSCLC and, in turn, increase the expression of anti-tumor microRNA let-7c. Also, let-7c directly targeted the 3′-untranslated regions of JMJD1A and EZH2. Taken together, JMJD1A could promote NSCLC tumorigenesis. JMJD1A/EZH2/let-7c constituted a feedback loop and might represent a promising therapeutic target for NSCLC.






Similar content being viewed by others
References
Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ. Cancer statistics, 2007. CA Cancer J Clin. 2007;57:43–66.
Jemal A, Siegel R, Ward E, Hao Y, Xu J, Murray T, et al. Cancer statistics, 2008. CA Cancer J Clin. 2008;58:71–96.
Shi Y, Lan F, Matson C, Mulligan P, Whetstine JR, Cole PA, et al. Histone demethylation mediated by the nuclear amine oxidase homolog lsd1. Cell. 2004;119:941–53.
Tsukada Y, Fang J, Erdjument-Bromage H, Warren ME, Borchers CH, Tempst P, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–6.
Tateishi K, Okada Y, Kallin EM, Zhang Y. Role of Jhdm2a in regulating metabolic gene expression and obesity resistance. Nature. 2009;458:757–61.
Okada Y, Scott G, Ray MK, Mishina Y, Zhang Y. Histone demethylase JHDM2A is critical for Tnp1 and Prm1 transcription and spermatogenesis. Nature. 2007;450:119–23.
Yamane K, Toumazou C, Tsukada Y, Erdjument-Bromage H, Tempst P, Wong J, et al. JHDM2A, a JmjC-containing H3K9 demethylase, facilitates transcription activation by androgen receptor. Cell. 2006;125:483–95.
Suikki HE, Kujala PM, Tammela TL, van Weerden WM, Vessella RL, Visakorpi T. Genetic alterations and changes in expression of histone demethylases in prostate cancer. Prostate. 2010;70:889–98.
Guo X, Shi M, Sun L, Wang Y, Gui Y, Cai Z, et al. The expression of histone demethylase JMJD1A in renal cell carcinoma. Neoplasma. 2011;58:153–7.
Cho HS, Toyokawa G, Daigo Y, Hayami S, Masuda K, Ikawa N, et al. The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int J Cancer. 2012;131:E179–189.
Yamada D, Kobayashi S, Yamamoto H, Tomimaru Y, Noda T, Uemura M, et al. Role of the hypoxia-related gene, JMJD1A, in hepatocellular carcinoma: clinical impact on recurrence after hepatic resection. Ann Surg Oncol. 2012;19 Suppl 3:S355–364.
Uemura M, Yamamoto H, Takemasa I, Mimori K, Hemmi H, Mizushima T, et al. Jumonji domain containing 1A is a novel prognostic marker for colorectal cancer: in vivo identification from hypoxic tumor cells. Clin Cancer Res Off J Am Assoc Cancer Res. 2010;16:4636–46.
Tee AE, Ling D, Nelson C, Atmadibrata B, Dinger ME, Xu N, Mizukami T, Liu PY, Liu B, Cheung B, Pasquier E, Haber M, Norris MD, Suzuki T, Marshall GM, Liu T. The histone demethylase JMJD1A induces cell migration and invasion by up-regulating the expression of the long noncoding RNA MALAT1. Oncotarget. 2014.
Fan L, Peng G, Sahgal N, Fazli L, Gleave M, Zhang Y, Hussain A, Qi J. Regulation of c-Myc expression by the histone demethylase JMJD1A is essential for prostate cancer cell growth and survival. Oncogene. 2015.
Hirsch FR, Varella-Garcia M, Bunn Jr PA, Di Maria MV, Veve R, Bremmes RM, et al. Epidermal growth factor receptor in non-small-cell lung carcinomas: correlation between gene copy number and protein expression and impact on prognosis. J Clin Oncol Off J Am Soc Clin Oncol. 2003;21:3798–807.
Marshall GM, Liu PY, Gherardi S, Scarlett CJ, Bedalov A, Xu N, et al. SIRT1 promotes N-Myc oncogenesis through a positive feedback loop involving the effects of MKP3 and ERK on N-Myc protein stability. PLoS Genet. 2011;7:e1002135.
Fussbroich B, Wagener N, Macher-Goeppinger S, Benner A, Falth M, Sultmann H, et al. EZH2 depletion blocks the proliferation of colon cancer cells. PLoS One. 2011;6:e21651.
Liu T, Tee AE, Porro A, Smith SA, Dwarte T, Liu PY, et al. Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for Myc oncogenesis. Proc Natl Acad Sci U S A. 2007;104:18682–7.
Ma X, Li C, Sun L, Huang D, Li T, He X, et al. Lin28/let-7 axis regulates aerobic glycolysis and cancer progression via PDK1. Nat Commun. 2014;5:5212.
Steele JC, Torr EE, Noakes KL, Kalk E, Moss PA, Reynolds GM, et al. The polycomb group proteins, BMI-1 and EZH2, are tumour-associated antigens. Br J Cancer. 2006;95:1202–11.
Piskounova E, Polytarchou C, Thornton JE, LaPierre RJ, Pothoulakis C, Hagan JP, et al. Lin28a and lin28b inhibit let-7 microRNA biogenesis by distinct mechanisms. Cell. 2011;147:1066–79.
Hayashi Y, Tsujii M, Wang J, Kondo J, Akasaka T, Jin Y, et al. CagA mediates epigenetic regulation to attenuate let-7 expression in Helicobacter pylori-related carcinogenesis. Gut. 2013;62:1536–46.
Sun D, Layer R, Mueller AC, Cichewicz MA, Negishi M, Paschal BM, et al. Regulation of several androgen-induced genes through the repression of the miR-99a/let-7c/miR-125b-2 miRNA cluster in prostate cancer cells. Oncogene. 2014;33:1448–57.
Zhang W, Liu H, Liu W, Liu Y, Xu J. Polycomb-mediated loss of microRNA let-7c determines inflammatory macrophage polarization via PAK1-dependent NF-κB pathway. Cell Death Differ. 2015;22:287–97.
Tzatsos A, Paskaleva P, Lymperi S, Contino G, Stoykova S, Chen Z, et al. Lysine-specific demethylase 2b (KDM2B)-let-7-enhancer of zester homolog 2 (EZH2) pathway regulates cell cycle progression and senescence in primary cells. J Biol Chem. 2011;286:33061–9.
Kong D, Heath E, Chen W, Cher ML, Powell I, Heilbrun L, et al. Loss of let-7 up-regulates EZH2 in prostate cancer consistent with the acquisition of cancer stem cell signatures that are attenuated by BR-DIM. PLoS One. 2012;7:e33729.
Wade MA, Jones D, Wilson L, Stockley J, Coffey K, Robson CN, et al. The histone demethylase enzyme KDM3A is a key estrogen receptor regulator in breast cancer. Nucleic Acids Res. 2015;43:196–207.
Parrish JK, Sechler M, Winn RA, Jedlicka P. The histone demethylase KDM3A is a microRNA-22-regulated tumor promoter in Ewing sarcoma. Oncogene. 2015;34:257–62.
Osawa T, Tsuchida R, Muramatsu M, Shimamura T, Wang F, Suehiro J, et al. Inhibition of histone demethylase JMJD1A improves anti-angiogenic therapy and reduces tumor-associated macrophages. Cancer Res. 2013;73:3019–28.
Krieg AJ, Rankin EB, Chan D, Razorenova O, Fernandez S, Giaccia AJ. Regulation of the histone demethylase JMJD1A by hypoxia-inducible factor 1 alpha enhances hypoxic gene expression and tumor growth. Mol Cell Biol. 2010;30:344–53.
Au SL, Wong CC, Lee JM, Fan DN, Tsang FH, Ng IO, et al. Enhancer of zeste homolog 2 epigenetically silences multiple tumor suppressor microRNAs to promote liver cancer metastasis. Hepatology. 2012;56:622–31.
Fujii S, Tokita K, Wada N, Ito K, Yamauchi C, Ito Y, et al. MEK-ERK pathway regulates EZH2 overexpression in association with aggressive breast cancer subtypes. Oncogene. 2011;30:4118–28.
Lu C, Han HD, Mangala LS, Ali-Fehmi R, Newton CS, Ozbun L, et al. Regulation of tumor angiogenesis by EZH2. Cancer Cell. 2010;18:185–97.
Behrens C, Solis LM, Lin H, Yuan P, Tang X, Kadara H, et al. Wistuba, II: EZH2 protein expression associates with the early pathogenesis, tumor progression, and prognosis of non-small cell lung carcinoma. Clin Cancer Res Off J Am Assoc Cancer Res. 2013;19:6556–65.
Liu L, Xu Z, Zhong L, Wang H, Jiang S, Long Q, et al. Prognostic value of EZH2 expression and activity in renal cell carcinoma: a prospective study. PLoS One. 2013;8:e81484.
Knudsen ES, Dervishaj O, Kleer CG, Pajak T, Schwartz GF, Witkiewicz AK. EZH2 and aldh1 expression in ductal carcinoma in situ: complex association with recurrence and progression to invasive breast cancer. Cell Cycle. 2013;12:2042–50.
Wang X, Zhao H, Lv L, Bao L, Wang X, Han S. Prognostic significance of EZH2 expression in non-small cell lung cancer: a meta-analysis. Sci Rep. 2016;6:19239.
Li Z, Xu L, Tang N, Xu Y, Ye X, Shen S, et al. The polycomb group protein EZH2 inhibits lung cancer cell growth by repressing the transcription factor Nrf2. FEBS Lett. 2014;588:3000–7.
Xu C, Hou Z, Zhan P, Zhao W, Chang C, Zou J, et al. EZH2 regulates cancer cell migration through repressing TIMP-3 in non-small cell lung cancer. Med Oncol. 2013;30:713.
Sun M, Liu XH, Lu KH, Nie FQ, Xia R, Kong R, et al. EZH2-mediated epigenetic suppression of long noncoding RNA SPRY4-IT1 promotes NSCLC cell proliferation and metastasis by affecting the epithelial-mesenchymal transition. Cell Death Dis. 2014;5:e1298.
Nadiminty N, Tummala R, Lou W, Zhu Y, Shi XB, Zou JX, et al. MicroRNA let-7c is downregulated in prostate cancer and suppresses prostate cancer growth. PLoS One. 2012;7:e32832.
Han HB, Gu J, Zuo HJ, Chen ZG, Zhao W, Li M, et al. Let-7c functions as a metastasis suppressor by targeting MMP11 and PBX3 in colorectal cancer. J Pathol. 2012;226:544–55.
Zhu XM, Wu LJ, Xu J, Yang R, Wu FS. Let-7c microRNA expression and clinical significance in hepatocellular carcinoma. J Int Med Res. 2011;39:2323–9.
Motoyama K, Inoue H, Nakamura Y, Uetake H, Sugihara K, Mori M. Clinical significance of high mobility group a2 in human gastric cancer and its relationship to let-7 microRNA family. Clin Cancer Res Off J Am Assoc Cancer Res. 2008;14:2334–40.
Takamizawa J, Konishi H, Yanagisawa K, Tomida S, Osada H, Endoh H, et al. Reduced expression of the let-7 microRNAs in human lung cancers in association with shortened postoperative survival. Cancer Res. 2004;64:3753–6.
Navarro A, Marrades RM, Vinolas N, Quera A, Agusti C, Huerta A, et al. MicroRNAs expressed during lung cancer development are expressed in human pseudoglandular lung embryogenesis. Oncology. 2009;76:162–9.
Nagayama K, Kohno T, Sato M, Arai Y, Minna JD, Yokota J. Homozygous deletion scanning of the lung cancer genome at a 100-kb resolution. Genes Chromosomes Cancer. 2007;46:1000–10.
Wang PY, Sun YX, Zhang S, Pang M, Zhang HH, Gao SY, et al. Let-7c inhibits A549 cell proliferation through oncogenic TRIB2 related factors. FEBS Lett. 2013;587:2675–81.
Zhan M, Qu Q, Wang G, Liu YZ, Tan SL, Lou XY, et al. Let-7c inhibits NSCLC cell proliferation by targeting HOXA1. Asian Pac J Cancer Prev. 2013;14:387–92.
Zhao B, Han H, Chen J, Zhang Z, Li S, Fang F, et al. MicroRNA let-7c inhibits migration and invasion of human non-small cell lung cancer by targeting ITGB3 and MAP4K3. Cancer Lett. 2014;342:43–51.
Wang H, Zhao Q, Deng K, Guo X, Xia J. Lin28: an emerging important oncogene connecting several aspects of cancer. Tumour Biol J Int Soc Oncodev Biol Med. 2016.
Zhang WC, Shyh-Chang N, Yang H, Rai A, Umashankar S, Ma S, et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell. 2012;148:259–72.
Zhou J, Ng SB, Chng WJ. Lin28/lin28b: an emerging oncogenic driver in cancer stem cells. Int J Biochem Cell Biol. 2013;45:973–8.
Acknowledgments
This work was supported by the 863 Project (No. 2012AA02A517), Medical Scientific Research Foundation of Guangdong Province (No. B2014362), and National Natural Science Foundation of China (No. 81260337).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None
Rights and permissions
About this article
Cite this article
Zhan, M., Wen, F., Liu, L. et al. JMJD1A promotes tumorigenesis and forms a feedback loop with EZH2/let-7c in NSCLC cells. Tumor Biol. 37, 11237–11247 (2016). https://doi.org/10.1007/s13277-016-4999-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-4999-9