Abstract
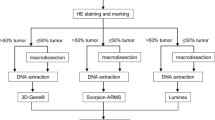
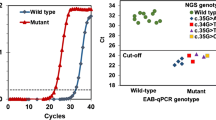
A gapped ligase chain reaction (gLCR)-based technique was developed and tested on clinical formalin-fixed, paraffin-embedded (FFPE) tissues from colorectal cancer patients. The technique was designed to detect low-level KRAS codon 12 or 13 mutations or confirming doubtful results gained by less sensitive KRAS screening techniques. The gLCR approach was compared with mutation screening techniques commonly used in routine diagnostics regarding sensitivity and specificity. The herein described monoplex gLCR technique is a useful and powerful tool for detecting low-level KRAS codon 12 and 13 mutations in a vast majority of wild type DNA. The gLCR has the capacity to detect one mutated allele in an excess of at least one million wild type alleles (0.0001 %) and is therefore an ideal technique for confirming doubtful KRAS mutation screening results obtained by other techniques. The variance of the gLCRs signal amplitude was very low and is showing a high reproducibility with constant sensitivity even at higher dilutions. The financial effort and the handling time for this technique are low and comparable to a standard cycle sequencing reaction. Additionally, the gLCR technique is easy extendable for the detection of many other clinical relevant mutation hotspots.


Similar content being viewed by others
References
Bogaert J, Prenen H. Molecular genetics of colorectal cancer. Ann Gastroenterol. 2014;27(1):9–14.
Brenner H, Hoffmeister M, Stegmaier C, Brenner G, Altenhofen L, Haug U. Risk of progression of advanced adenomas to colorectal cancer by age and sex: estimates based on 840,149 screening colonoscopies. Gut. 2007;56(11):1585–9.
Vogelstein B, Fearon ER, Hamilton SR, Kern SE, Preisinger AC, Leppert M, et al. Genetic alterations during colorectal-tumor development. N Engl J Med. 1988;319(9):525–32.
Boland CR, Shin SK, Goel A. Promoter methylation in the genesis of gastrointestinal cancer. Yonsei Med J. 2009;50(3):309–21.
Bohanes P, LaBonte MJ, Winder T, Lenz HJ. Predictive molecular classifiers in colorectal cancer. Semin Oncol. 2011;38(4):576–87.
Wheeler DL, Dunn EF, Harari PM. Understanding resistance to EGFR inhibitors-impact on future treatment strategies. Nat Rev Clin Oncol. 2010;7(9):493–507.
Bibeau F, Boissière-Michot F, Sabourin JC, Gourgou-Bourgade S, Radal M, Penault-Llorca F, et al. Assessment of epidermal growth factor receptor (EGFR) expression in primary colorectal carcinomas and their related metastases on tissue sections and tissue microarray. Virchows Arch. 2006;449(3):281–7.
Van Cutsem E, Köhne CH, Hitre E, Zaluski J, Chang Chien CR, Makhson A, et al. Cetuximab and chemotherapy as initial treatment for metastatic colorectal cancer. N Engl J Med. 2009;360(14):1408–17.
Kim K. Early detection and prevention of colorectal cancer. New Jersey: SLACK Incorporated MEDICAL/Gastroenterology; 2009.
Wiedmann M, Wilson WJ, Czajka J, Luo J, Barany F, Batt CA. Ligase chain reaction (LCR)—overview and applications. PCR Methods Appl. 1994;3(4):S51–64.
Harden SV, Thomas DC, Benoit N, Minhas K, Westra WH, Califano JA, et al. Real-time gap ligase chain reaction: a rapid semiquantitative assay for detecting p53 mutation at low levels in surgical margins and lymph nodes from resected lung and head and neck tumors. Clin Cancer Res. 2004;10(7):2379–85.
Song Y, Zhang Y, Wang TH. Single quantum dot analysis enables multiplexed point mutation detection by gap ligase chain reaction. Small. 2013;9(7):1096–105. doi:10.1002/smll.201202242.
Landegren U, Kaiser R, Sanders J, Hood L. A ligase-mediated gene detection technique. Science. 1988;241(4869):1077–80.
Sauer S (2001) Technology development for genotyping single nucleotide polymorphisms. Dissertation, Freie Universität Berlin
Khanna M, Park P, Zirvi M, Cao W, Picon A, Day J, et al. Multiplex PCR/LDR for detection of K-ras mutations in primary colon tumors. Oncogene. 1999;18(1):27–38.
Ginya H, Matsushita R, Yohda M. Quantification and improvement of error rate during ligase detection reaction. J Biosci Bioeng. 2010;109(2):202–4.
Abravaya K, Carrino JJ, Muldoon S, Lee HH. Detection of point mutations with a modified ligase chain reaction (Gap-LCR). Nucleic Acids Res. 1995;23(4):675–82.
Hahn M, Wilhelm J, Pingoud A. Influence of fluorophor dye labels on the migration behavior of polymerase chain reaction—amplified short tandem repeats during denaturing capillary electrophoresis. Electrophoresis. 2001;22(13):2691–700.
Davidson CJ, Zeringer E, Champion KJ, Gauthier MP, Wang F, Boonyaratanakornkit J, et al. Improving the limit of detection for Sanger sequencing: a comparison of methodologies for KRAS variant detection. Biotechniques. 2012;14:2012. doi:10.2144/000113913.
Park JH, Kim IJ, Kang HC, Shin Y, Park HW, Jang SG, et al. Oligonucleotide microarray-based mutation detection of the K-ras gene in colorectal cancers with use of competitive DNA hybridization. Clin Chem. 2004;50(9):1688–91.
Yi P, Chen Z, Yu L, Zheng Y, Liu G, Xie H, et al. Development of a PCR/LDR/capillary electrophoresis assay with potential for the detection of a beta-thalassemia fetal mutation in maternal plasma. J Matern Fetal Neonatal Med. 2010;23(8):920–7.
Weichert W, Schewe C, Lehmann A, Sers C, Denkert C, Budczies J, et al. KRAS genotyping of paraffin-embedded colorectal cancer tissue in routine diagnostics: comparison of methods and impact of histology. J Mol Diagn. 2010;12(1):35–42.
Acknowledgments
This research was supported by a grant of Prof. Dr. med. A. Bosse, head of the Division of Molecular Pathology, Department of Pathology, Katharinen Hospital Stuttgart, Germany. We also thank Dr. med. K. H. Wiedorn for supporting this study as an expert pathologist in all medical aspects.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 393 kb)
Rights and permissions
About this article
Cite this article
Jenner, S., Techel, D. Development of a gLCR-based KRAS mutation detection approach and its comparison with other screening methods. Tumor Biol. 36, 6361–6368 (2015). https://doi.org/10.1007/s13277-015-3323-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3323-4