Abstract
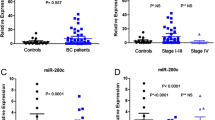
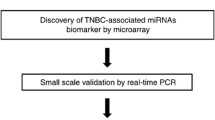
Cancer-associated microRNAs have been stably detected in blood. The objective of this study was to identify a panel of circulating microRNAs with the potential to serve as biomarkers for estrogen receptor-positive (ER+)/human epidermal growth factor receptor 2 (HER2)− breast cancer. We used microarray-based expression profiling to compare the levels of circulating microRNAs in blood samples from 11 ER+/HER2− advanced breast cancer patients plus 5 age-matched controls. MicroRNA levels were validated by reverse transcription quantitative polymerase chain reaction in 40 control subjects, 187 early breast cancer patients, and 45 metastatic breast cancer patients. Then, we assessed the association between the levels of microRNA and clinical outcomes of ER+/HER2− metastatic breast cancer. Initially, we found that miR-1280, miR-1260, and miR-720 were up-regulated in blood from breast cancer patients (P < 0.05). In validation, miR-1280 levels significantly increased in breast cancer patients and reflected tumor status (control<<early cancer<metastatic cancer). Among 37 metastatic breast cancer patients, miR-1280 levels significantly decreased after treatment in patients who responded to systemic treatment (P < 0.001). We confirmed that miR-1280 was not a classic microRNA, but a tRNALeu-derived fragment. These findings suggest that a circulating tRNA-derived microRNA, miR-1280, is differently expressed in breast cancer patients and may serve as a biomarker for ER-positive breast cancer.





Similar content being viewed by others
Abbreviations
- ER:
-
Estrogen receptor
- HER2:
-
Human epidermal growth factor receptor 2
- EBC:
-
Early breast cancer
- MBC:
-
Metastatic breast cancer
- PCR:
-
Polymerase chain reaction
- qRT-PCR:
-
Quantitative reverse transcription PCR
- DIG:
-
Digoxigenin
- SD:
-
Standard deviation
- RECIST:
-
Response Evaluation Criteria In Solid Tumors
- OS:
-
Overall survival
- RPA1:
-
Replication protein A1
References
Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69.
Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66.
Iorio M, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–70.
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435(7043):834–8.
Garzon R, Calin G, Croce C. MicroRNAs in cancer. Annu Rev Med. 2009;60:167–79.
Garzon R, Marcucci G, Croce C. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9(10):775–89.
Kluiver J, Poppema S, de Jong D, Blokzijl T, Harms G, Jacobs S, et al. BIC and miR-155 are highly expressed in Hodgkin, primary mediastinal and diffuse large B cell lymphomas. J Pathol. 2005;207(2):243–9.
Chan J, Krichevsky A, Kosik K. MicroRNA-21 is an antiapoptotic factor in human glioblastoma cells. Cancer Res. 2005;65(14):6029–33.
Ventura A, Young A, Winslow M, Lintault L, Meissner A, Erkeland S, et al. Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell. 2008;132(5):875–86.
Lowery A, Miller N, Devaney A, McNeill RE, Davoren PA, Lemetre C, et al. MicroRNA signatures predict oestrogen receptor, progesterone receptor and HER2/neu receptor status in breast cancer. Breast Cancer Res. 2009;11(3):R27.
Blenkiron C, Goldstein L, Thorne N, Spiteri I, Chin S-F, Dunning M, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214.
Lee DY, Deng Z, Wang CH, Yang BB. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc Natl Acad Sci U S A. 2007;104(51):20350–5.
Shimono Y, Zabala M, Cho R, Lobo N, Dalerba P, Qian D, et al. Downregulation of miRNA-200c links breast cancer stem cells with normal stem cells. Cell. 2009;138(3):592–603.
Shenouda S, Alahari S. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3–4):369–78.
Mitchell P, Parkin R, Kroh E, Fritz B, Wyman S, Pogosova-Agadjanyan E, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8.
Heneghan H, Miller N, Lowery A. Circulating microRNAs as novel minimally invasive biomarkers for breast cancer. Ann Surg. 2010;251(3):499–505.
Tsujiura M, Ichikawa D, Komatsu S, Shiozaki A, Takeshita H, Kosuga T, et al. Circulating microRNAs in plasma of patients with gastric cancers. Br J Cancer. 2010;102(7):1174–9.
Wong T-S, Liu X-B, Wong B. Mature miR-184 as potential oncogenic microRNA of squamous cell carcinoma of tongue. Clin Cancer Res. 2008;14(9):2588–92.
Hu Z, Chen X, Zhao Y, Tian T, Jin G, Shu Y, et al. Serum microRNA signatures identified in a genome-wide serum microRNA expression profiling predict survival of non-small-cell lung cancer. J Clin Oncol. 2010;28(10):1721–6.
Sun Y, Wang M, Lin G, Sun S, Li X, Qi J, et al. Serum microRNA-155 as a potential biomarker to track disease in breast cancer. PLoS One. 2012;7(10):e47003.
Zhao H, Shen J, Medico L, Wang D, Ambrosone C. A pilot study of circulating miRNAs as potential biomarkers of early stage breast cancer. PLoS One. 2010;5(10):e13735.
Comparison of microRNA expression with whole blood between ER positive breast cancer patients and normal controls. http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE53179.
Schopman NC, Heynen S, Haasnoot J, Berkhout B. A miRNA-tRNA mix-up: tRNA origin of proposed miRNA. RNA Biol. 2010;7(5):573–6.
Cheloufi S, Dos Santos C, Chong M, Hannon G. A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature. 2010;465(7298):584–9.
Yang J, Lai E. Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell. 2011;43(6):892–903.
Lee Y, Shibata Y, Malhotra A, Dutta A. A novel class of small RNAs: tRNA-derived RNA fragments (tRFs). Genes Dev. 2009;23(22):2639–49.
Maute RL, Schneider C, Sumazin P, Holmes A, Califano A, Basso K, et al. tRNA-derived microRNA modulates proliferation and the DNA damage response and is down-regulated in B cell lymphoma. Proc Natl Acad Sci U S A. 2013;110(4):1404–9.
Chan M, Liaw C, Ji S, Tan H, Wong C, Thike A, et al. Identification of circulating microRNA signatures for breast cancer detection. Clin Cancer Res. 2013;19(16):4477–87.
Jung E-J, Santarpia L, Kim J, Esteva F, Moretti E, Buzdar A, et al. Plasma microRNA 210 levels correlate with sensitivity to trastuzumab and tumor presence in breast cancer patients. Cancer. 2012;118(10):2603–14.
Acknowledgments
This work was supported by NCC Grant 0910220 and NCC Grant 1110260.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Park, I.H., Kang, J.H., Lee, K.S. et al. Identification and clinical implications of circulating microRNAs for estrogen receptor-positive breast cancer. Tumor Biol. 35, 12173–12180 (2014). https://doi.org/10.1007/s13277-014-2525-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-014-2525-5