Abstract
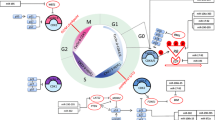
MicroRNAs (miRNAs) are small noncoding RNAs of 19–25 nt that can regulate gene expression at a posttranscriptional level. Increasing evidence indicates that miRNAs participate in almost every step of cellular processes and are often aberrantly expressed in human cancer. miR-221 and miR-222 are two highly homologous miRNAs that always act as a gene cluster (miR-221/222) in cellular regulation and have extensively been studied in cancer network. Here, we review the role of miR-221/222 in breast cancer (BCa) development and progression: regulating proliferative signaling pathways, altering telomere and telomerase activity, avoiding cell death from tumor suppressors, autophagy and apoptosis, monitoring angiogenesis, supporting epithelial–mesenchymal transition, and even controlling cell-specific function within microenvironment. We consider that miR-221/222 act as promising biomarkers for BCa and they would offer a new way in molecular targeting cancer treatment.




Similar content being viewed by others
References
Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33.
Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–58.
O'Day E, Lal A. MicroRNAs and their target gene networks in breast cancer. Breast Cancer Res. 2010;12(2):201.
Blenkiron C, Goldstein LD, Thorne NP, Spiteri I, Chin SF, Dunning MJ, et al. MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol. 2007;8(10):R214.
Garofalo M, Quintavalle C, Romano G, Croce CM, Condorelli G. miR221/222 in cancer: their role in tumor progression and response to therapy. Curr Mol Med. 2012;12(1):27–33.
Howe EN, Cochrane DR, Richer JK. The miR-200 and miR-221/222 microRNA families: opposing effects on epithelial identity. J Mammary Gland Biol Neoplasia. 2012;17(1):65–77.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–74.
Miller TE, Ghoshal K, Ramaswamy B, Roy S, Datta J, Shapiro CL, et al. MicroRNA-221/222 confers tamoxifen resistance in breast cancer by targeting p27Kip1. J Biol Chem. 2008;283(44):29897–903.
Zhao JJ, Lin J, Yang H, Kong W, He L, Ma X, et al. MicroRNA-221/222 negatively regulates estrogen receptor alpha and is associated with tamoxifen resistance in breast cancer. J Biol Chem. 2008;283(45):31079–86.
Cochrane DR, Cittelly DM, Howe EN, Spoelstra NS, McKinsey EL, LaPara K, et al. MicroRNAs link estrogen receptor alpha status and Dicer levels in breast cancer. Horm Cancer. 2010;1(6):306–19.
Zhao R, Wu J, Jia W, Gong C, Yu F, Ren Z, et al. Plasma miR-221 as a predictive biomarker for chemoresistance in breast cancer patients who previously received neoadjuvant chemotherapy. Onkologie. 2011;34(12):675–80.
García-Becerra R, Santos N, Díaz L, Camacho J. Mechanisms of resistance to endocrine therapy in breast cancer: focus on signaling pathways, miRNAs and genetically based resistance. Int J Mol Sci. 2012;14(1):108–45.
Manavalan TT, Teng Y, Appana SN, Datta S, Kalbfleisch TS, Li Y, et al. Differential expression of microRNA expression in tamoxifen-sensitive MCF-7 versus tamoxifen-resistant LY2 human breast cancer cells. Cancer Lett. 2011;313(1):26–43.
Lu Y, Roy S, Nuovo G, Ramaswamy B, Miller T, Shapiro C, et al. Anti-microRNA-222 (anti-miR-222) and -181B suppress growth of tamoxifen-resistant xenografts in mouse by targeting TIMP3 protein and modulating mitogenic signal. J Biol Chem. 2011;286(49):42292–302.
Di Leva G, Gasparini P, Piovan C, Ngankeu A, Garofalo M, Taccioli C, et al. MicroRNA cluster 221–222 and estrogen receptor alpha interactions in breast cancer. J Natl Cancer Inst. 2010;102(10):706–21.
McCafferty MP, McNeill RE, Miller N, Kerin MJ. Interactions between the estrogen receptor, its cofactors and microRNAs in breast cancer. Breast Cancer Res Treat. 2009;116(3):425–32.
Rao X, Di Leva G, Li M, Fang F, Devlin C, Hartman-Frey C, et al. MicroRNA-221/222 confers breast cancer fulvestrant resistance by regulating multiple signaling pathways. Oncogene. 2011;30(9):1082–97.
Yoshimoto N, Toyama T, Takahashi S, Sugiura H, Endo Y, Iwasa M, et al. Distinct expressions of microRNAs that directly target estrogen receptor α in human breast cancer. Breast Cancer Res Treat. 2011;130(1):331–9.
Leung YK, Lee MT, Lam HM, Tarapore P, Ho SM. Estrogen receptor-beta and breast cancer: translating biology into clinical practice. Steroids. 2012;77(7):727–37.
Wärnmark A, Almlöf T, Leers J, Gustafsson JA, Treuter E. Differential recruitment of the mammalian mediator subunit TRAP220 by estrogen receptors ERalpha and ERbeta. J Biol Chem. 2001;276(26):23397–404.
Yang Z, Barnes CJ, Kumar R. Human epidermal growth factor receptor 2 status modulates subcellular localization of and interaction with estrogen receptor alpha in breast cancer cells. Clin Cancer Res. 2004;10(11):3621–8.
Stinson S, Lackner MR, Adai AT, Yu N, Kim HJ, O'Brien C, et al. TRPS1 targeting by miR-221/222 promotes the epithelial-to-mesenchymal transition in breast cancer. Sci Signal. 2011;4(177):ra41.
Cui J, Germer K, Wu T, Wang J, Luo J, Wang SC, et al. Cross-talk between HER2 and MED1 regulates tamoxifen resistance of human breast cancer cells. Cancer Res. 2012;72(21):5625–34.
Vargas J, Feltes BC, Poloni Jde F, Lenz G, Bonatto D. Senescence: an endogenous anticancer mechanism. Front Biosci. 2012;17:2616–43.
Nabetani A, Ishikawa F. Alternative lengthening of telomeres pathway: recombination-mediated telomere maintenance mechanism in human cells. J Biochem. 2011;149(1):5–14.
Cheung AL, Deng W. Telomere dysfunction, genome instability and cancer. Front Biosci. 2008;13:2075–90.
Benetti R, Gonzalo S, Jaco I, Muñoz P, Gonzalez S, Schoeftner S, et al. A mammalian microRNA cluster controls DNA methylation and telomere recombination via Rbl2-dependent regulation of DNA methyltransferases. Nat Struct Mol Biol. 2008;15(3):268–79.
Romilda C, Marika P, Alessandro S, Enrico L, Marina B, Andromachi K, et al. Oxidative DNA damage correlates with cell immortalization and miR-92 expression in hepatocellular carcinoma. BMC Cancer. 2012;12:177.
Cairney CJ, Keith WN. Telomerase redefined: integrated regulation of hTR and hTERT for telomere maintenance and telomerase activity. Biochimie. 2008;90(1):13–23.
Mitomo S, Maesawa C, Ogasawara S, Iwaya T, Shibazaki M, Yashima-Abo A, et al. Downregulation of miR-138 is associated with overexpression of human telomerase reverse transcriptase protein in human anaplastic thyroid carcinoma cell lines. Cancer Sci. 2008;99(2):280–6.
Lu L, Zhang C, Zhu G, Irwin M, Risch H, Menato G, et al. Telomerase expression and telomere length in breast cancer and their associations with adjuvant treatment and disease outcome. Breast Cancer Res. 2011;13(3):R56.
Winnikow EP, Medeiros LR, Edelweiss MI, Rosa DD, Edelweiss M, Simões PW, et al. Accuracy of telomerase in estimating breast cancer risk: a systematic review and meta-analysis. Breast. 2012;21(1):1–7.
Shen J, Gammon MD, Terry MB, Bradshaw PT, Wang Q, Teitelbaum SL, et al. Genetic polymorphisms in telomere pathway genes, telomere length, and breast cancer survival. Breast Cancer Res Treat. 2012;134(1):393–400.
Lu X, Zhao P, Zhang C, Fu Z, Chen Y, Lu A, et al. Analysis of miR-221 and p27 expression in human gliomas. Mol Med Rep. 2009;2(4):651–6.
Pineau P, Volinia S, McJunkin K, Marchio A, Battiston C, Terris B, et al. miR-221 overexpression contributes to liver tumorigenesis. Proc Natl Acad Sci. 2010;107(1):264–9.
Galardi S, Mercatelli N, Giorda E, Massalini S, Frajese GV, Ciafrè SA, et al. miR-221 and miR-222 expression affects the proliferation potential of human prostate carcinoma cell lines by targeting p27Kip1. J Biol Chem. 2007;282(32):23716–24.
Kedde M, van Kouwenhove M, Zwart W, Oude Vrielink JA, Elkon R, Agami RA. Pumilio-induced RNA structure switch in p27-3′ UTR controls miR-221 and miR-222 accessibility. Nat Cell Biol. 2010;12(10):1014–20.
Garofalo M, Di Leva G, Romano G, Nuovo G, Suh SS, Ngankeu A, et al. miR-221&222 regulate TRAIL resistance and enhance tumorigenicity through PTEN and TIMP3 downregulation. Cancer Cell. 2009;16(6):498–509.
Chen Y, Zaman MS, Deng G, Majid S, Saini S, Liu J, et al. MicroRNAs 221/222 and genistein-mediated regulation of ARHI tumor suppressor gene in prostate cancer. Cancer Prev Res (Phila). 2011;4(1):76–86.
Vousden KH, Prives C. Blinded by the light: the growing complexity of p53. Cell. 2009;137(3):413–31.
Frankel LB, Lund AH. MicroRNA regulation of autophagy. Carcinogenesis. 2012;33(11):2018–25.
Pyo JO, Nah J, Jung YK. Molecules and their functions in autophagy. Exp Mol Med. 2012;44(2):73–80.
Noren Hooten N, Abdelmohsen K, Gorospe M, Ejiogu N, Zonderman AB, Evans MK. microRNA expression patterns reveal differential expression of target genes with age. PLoS One. 2010;5(5):e10724.
Gramantieri L, Fornari F, Ferracin M, Veronese A, Sabbioni S, Calin GA, et al. MicroRNA-221 targets Bmf in hepatocellular carcinoma and correlates with tumor multifocality. Clin Cancer Res. 2009;15(16):5073–81.
Zhang C, Zhang J, Zhang A, Wang Y, Han L, You Y, et al. PUMA is a novel target of miR-221/222 in human epithelial cancers. Int J Oncol. 2010;37(6):1621–6.
Guttilla IK, Phoenix KN, Hong X, Tirnauer JS, Claffey KP, White BA. Prolonged mammosphere culture of MCF-7 cells induces an EMT and repression of the estrogen receptor by microRNAs. Breast Cancer Res Treat. 2012;132(1):75–85.
Aschrafi A, Schwechter AD, Mameza MG, Natera-Naranjo O, Gioio AE, Kaplan BB. MicroRNA-338 regulates local cytochrome c oxidase IV mRNA levels and oxidative phosphorylation in the axons of sympathetic neurons. J Neurosci. 2008;28(47):12581–90.
Gong M, Chen Y, Senturia R, Ulgherait M, Faller M, Guo F. Caspases cleave and inhibit the microRNA processing protein DiGeorge Critical Region 8. Protein Sci. 2012;21(6):797–808.
Urbich C, Kuehbacher A, Dimmeler S. Role of microRNAs in vascular diseases, inflammation, and angiogenesis. Cardiovasc Res. 2008;79(4):581–8.
Rippe C, Blimline M, Magerko KA, Lawson BR, LaRocca TJ, Donato AJ, et al. MicroRNA changes in human arterial endothelial cells with senescence: relation to apoptosis, eNOS and inflammation. Exp Gerontol. 2012;47(1):45–51.
Durán WN, Breslin JW, Sánchez FA. The NO cascade, eNOS location, and microvascular permeability. Cardiovasc Res. 2010;87(2):254–61.
Suárez Y, Fernández-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ Res. 2007;100(8):1164–73.
Chen Y, Banda M, Speyer CL, Smith JS, Rabson AB, Gorski DH. Regulation of the expression and activity of the antiangiogenic homeobox gene GAX/MEOX2 by ZEB2 and microRNA-221. Mol Cell Biol. 2010;30(15):3902–13.
Liu X, Cheng Y, Yang J, Xu L, Zhang C. Cell-specific effects of miR-221/222 in vessels: molecular mechanism and therapeutic application. J Mol Cell Cardiol. 2012;52(1):245–55.
Liu X, Cheng Y, Zhang S, Lin Y, Yang J, Zhang C. A necessary role of miR-221 and miR-222 in vascular smooth muscle cell proliferation and neointimal hyperplasia. Circ Res. 2009;104(4):476–87.
Poliseno L, Tuccoli A, Mariani L, Evangelista M, Citti L, Woods K, et al. MicroRNAs modulate the angiogenic properties of HUVECs. Blood. 2006;108(9):3068–71.
Noonan DM, De Lerma BA, Vannini N, Mortara L, Albini A. Inflammation, inflammatory cells and angiogenesis: decisions and indecisions. Cancer Metastasis Rev. 2008;27(1):31–40.
Dentelli P, Rosso A, Orso F, Olgasi C, Taverna D, Brizzi MF. microRNA-222 controls neovascularization by regulating signal transducer and activator of transcription 5A expression. Arterioscler Thromb Vasc Biol. 2010;30(8):1562–8.
Zhu N, Zhang D, Chen S, Liu X, Lin L, Huang X, et al. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis. 2011;215(2):286–93.
Gao D, Vahdat LT, Wong S, Chang JC, Mittal V. Microenvironmental regulation of epithelial–mesenchymal transitions in cancer. Cancer Res. 2012;72(19):4883–9.
Shah MY, Calin GA. MicroRNAs miR-221 and miR-222: a new level of regulation in aggressive breast cancer. Genome Med. 2011;3(8):56.
Radojicic J, Zaravinos A, Vrekoussis T, Kafousi M, Spandidos DA, Stathopoulos EN. MicroRNA expression analysis in triple-negative (ER, PR and Her2/neu) breast cancer. Cell Cycle. 2011;10(3):507–17.
Wellner U, Schubert J, Burk UC, Schmalhofer O, Zhu F, Sonntag A, et al. The EMT-activator ZEB1 promotes tumorigenicity by repressing stemness-inhibiting microRNAs. Nat Cell Biol. 2009;11(12):1487–95.
de Herreros AG, Peiró S, Nassour M, Savagner P. Snail family regulation and epithelial mesenchymal transitions in breast cancer progression. J Mammary Gland Biol Neoplasia. 2010;15(2):135–47.
Sleeman JP, Thiery JP. SnapShot: the epithelial–mesenchymal transition. Cell. 2011;145(1):162.
Lambertini E, Lolli A, Vezzali F, Penolazzi L, Gambari R, Piva R. Correlation between Slug transcription factor and miR-221 in MDA-MB-231 breast cancer cells. BMC Cancer. 2012;12:445.
Ye Y, Xiao Y, Wang W, Yearsley K, Gao JX, Shetuni B, et al. ER alpha signaling through slug regulates E-cadherin and EMT. Oncogene. 2010;29(10):1451–62.
Grelier G, Voirin N, Ay AS, Cox DG, Chabaud S, Treilleux I, et al. Prognostic value of Dicer expression in human breast cancers and association with the mesenchymal phenotype. Br J Cancer. 2009;101(4):673–83.
Heldin CH, Vanlandewijck M, Moustakas A. Regulation of EMT by TGFβ in cancer. FEBS Lett. 2012;586(14):1959–70.
Mayoral RJ, Pipkin ME, Pachkov M, van Nimwegen E, Rao A, Monticelli S. MicroRNA-221-222 regulate the cell cycle in mast cells. J Immunol. 2009;182(1):433–45.
Mayoral RJ, Deho L, Rusca N, Bartonicek N, Saini HK, Enright AJ, et al. MiR-221 influences effector functions and actin cytoskeleton in mast cells. PLoS One. 2011;6(10):e26133.
Lu C, Huang X, Zhang X, Roensch K, Cao Q, Nakayama KI, et al. miR-221 and miR-155 regulate human dendritic cell development, apoptosis, and IL-12 production through targeting of p27kip1, KPC1, and SOCS-1. Blood. 2011;117(16):4293–303.
Zhao L, Sun Y, Hou Y, Peng Q, Wang L, Luo H, et al. MiRNA expression analysis of cancer-associated fibroblasts and normal fibroblasts in breast cancer. Int J Biochem Cell Biol. 2012;44(11):2051–9.
Brognara E, Fabbri E, Aimi F, Manicardi A, Bianchi N, Finotti A, et al. Peptide nucleic acids targeting miR-221 modulate p27Kip1 expression in breast cancer MDA-MB-231 cells. Int J Oncol. 2012;41(6):2119–27.
Kim JK, Choi KJ, Lee M, Jo MH, Kim S. Molecular imaging of a cancer-targeting theragnostics probe using a nucleolin aptamer- and microRNA-221 molecular beacon-conjugated nanoparticle. Biomaterials. 2012;33(1):207–17.
Acknowledgments
This work has no fund. We thank Shan-Liang Zhong, MD and Jin-Jin Xu, MD for their discussions and help in revision.
Conflicts of interest
None
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Chen, WX., Hu, Q., Qiu, MT. et al. miR-221/222: promising biomarkers for breast cancer. Tumor Biol. 34, 1361–1370 (2013). https://doi.org/10.1007/s13277-013-0750-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-013-0750-y