Abstract
Backgrounds
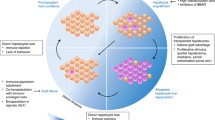
Orthotopic Liver Transplantation (OLT) is the therapy of choice for the treatment of end-stage liver disease, but the severe shortage of organ donors, complex and expensive surgical procedure and increased mortality of prospective organ recipient limit the use of OLT. To overcome this problem the technique of hepatocyte transplantation has been considered as an alternative to OLT. Hepatocyte transplantation is less invasive, cost effective cryo-preservable and can be distributed from single donor to multiple recipients. In this study we have established and characterize the hepatocyte cell line possessing the morphological and functional characteristics of hepatocytes from chemically injured liver. Hence we hypothesized that the hepatocyte cell line derived from liver injury can be used for hepatocyte transplantation.
Methods
To induce the priming of hepatocyte, mice were fed with 3,5-diethoxycarbonyl-1,4-dihydro-collidine (DDC) diet for 3 weeks and hepatocytes were obtained by two step collagenase perfusion method, hepatocytes hence obtained was expended by >300 passages and tested for various hepatocyte specific functions.
Results
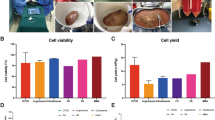
The cell line derived from liver injury model retains morphological and functional characteristics of hepatocytes in long-term in vitro culture. These cells transplanted to mice showed significant survival rate.
Conclusion
The Hepatocyte cell line derived from liver injury model were used for hepatocyte transplantation and showed the significantly higher survival rate.
Similar content being viewed by others
References
Byass, P. The global burden of liver disease: a challenge for methods and for public health. BMC Med 12, 159 (2014).
Reid, Y., Gaddipati, J. P., Yadav, D. & Kantor, J. Establishment of a human neonatal hepatocyte cell line. In Vitro Cell Dev Biol Anim 45, 535–542 (2009).
Katenz, E. et al. Cryopreservation of primary human hepatocytes: the benefit of trehalose as an additional cryoprotective agent. Liver Transpl 13, 38–45 (2007).
Hewitt, N. J. Optimisation of the cryopreservation of primary hepatocytes. Methods Mol Biol 640, 83–105 (2010).
Gramignoli, R. et al. Hypothermic storage of human hepatocytes for transplantation. Cell Transplant 23, 1143–1151 (2014).
Nagata, H. et al. Treatment of cirrhosis and liver failure in rats by hepatocyte xenotransplantation. Gastroenterology 124, 422–431 (2003).
Zarrinpar, A. & Busuttil, R. W. Liver transplantation: past, present and future. Nat Rev Gastroenterol Hepatol 10, 434–440 (2013).
Schumacher, I. K. et al. Transplantation of conditionally immortalized hepatocytes to treat hepatic encephalopathy. Hepatology 24, 337–343 (1996).
Vondran, F. W. et al. Isolation of primary human hepatocytes after partial hepatectomy: criteria for identification of the most promising liver specimen. Artificial Organs 32, 205–213 (2008).
Wiemann, S. U. et al. Hepatocyte telomere shortening and senescence are general markers of human liver cirrhosis. FASEB J 16, 935–942 (2002).
Chan, C. et al. Hepatic tissue engineering for adjunct and temporary liver support: critical technologies. Liver Transpl 10, 1331–1342 (2004).
Ramboer, E. et al. Strategies for immortalization of primary hepatocytes. J Hepatol 61, 925–943 (2014).
Risal, P. et al. The establishment and characterization of immortal hepatocyte cell lines from a mouse liver injury model. In Vitro Cell Dev Biol Anim 47, 526–534 (2011).
Mahieu-Caputo, D. et al. Repopulation of athymic mouse liver by cryopreserved early human fetal hepatoblasts. Hum Gene Ther 15, 1219–1228 (2004).
Farrugia, A. Albumin usage in clinical medicine: tradition or therapeutic? Transfus Med Rev 24, 53–63 (2010).
Takeichi, M. Functional correlation between cell adhesive properties and some cell surface proteins. J Cell Biol 75, 464–474 (1977).
Gumbiner, B. M. Regulation of cadherin-mediated adhesion in morphogenesis. Nat Rev Mol Cell Biol 6, 622–634 (2005).
Halbleib, J. M. & Nelson, W. J. Cadherins in development: cell adhesion, sorting, and tissue morphogenesis. Genes Dev 20, 3199–3214 (2006).
Lien, W. H., Klezovitch, O. & Vasioukhin, V. Cadherincatenin proteins in vertebrate development. Curr Opin Cell Biol 18, 499–506 (2006).
van Roy, F. & Berx, G. The cell-cell adhesion molecule E-cadherin. Cell Mol Life Sci 65, 3756–3788 (2008).
Tepass, U., Truong, K., Godt, D., Ikura, M. & Peifer, M. Cadherins in embryonic and neural morphogenesis. Nat Rev Mol Cell Biol 1, 91–100 (2000).
Krajewski, W. W. et al. Crystal structures of mammalian glutamine synthetases illustrate substrate-induced conformational changes and provide opportunities for drug and herbicide design. J Mol Biol 375, 217–228 (2008).
He, Y., Hakvoort, T. B., Vermeulen, J. L., Lamers, W. H. & Va. Roon, M. A. Glutamine synthetase is essential in early mouse embryogenesis. Dev Dyn 236, 1865–1875 (2007).
Harth, G., Maslesa-Galic, S., Tullius, M. V. & Horwitz, M. A. All four Mycobacterium tuberculosis glnA genes encode glutamine synthetase activities but only GlnA1 is abundantly expressed and essential for bacterial homeostasis. Mol Microbiol 58, 1157–1172 (2005).
Pan, X. et al. Establishment and characterization of immortalized human hepatocyte cell line for applications in bioartificial livers. Biotechnol Lett 34, 2183–2190 (2012).
Tomasi, T. B.. Jr. Structure and function of alpha-fetoprotein. Annu Rev Med 28, 453–465 (1977).
Ruoslahti, E., Pihko, H. & Seppala, M. Alpha-fetoprotein: immunochemical purification and chemical properties. Expression in normal state and in malignant and non-malignant liver disease. Transplant Rev 20, 38–60 (1974).
Jungermann, K. & Katz, N. Functional specialization of different hepatocyte populations. Physiol Rev 69, 708–764 (1989).
Costa, R. H., Kalinichenko, V. V., Holterman, A. X. & Wang, X. Transcription factors in liver development, differentiation, and regeneration. Hepatology 38, 1331–1347 (2003).
Mendel, D. B., Hansen, L. P., Graves, M. K., Conley, P. B. & Crabtree, G. R. HNF-1 alpha and HNF-1 beta (vHNF-1) share dimerization and homeo domains, but not activation domains, and form heterodimers in vitro. Genes Dev 5, 1042–1056 (1991).
Rausa, F. M., Tan, Y. & Costa, R. H. Association between hepatocyte nuclear factor 6 (HNF-6) and FoxA2 DNA binding domains stimulates FoxA2 transcriptional activity but inhibits HNF-6 DNA binding. Mol Cell Biol 23, 437–449 (2003).
Sakaguchi, T. et al. Cloning of the human claudin-2 5′-flanking region revealed a TATA-less promoter with conserved binding sites in mouse and human for caudalrelated homeodomain proteins and hepatocyte nuclear factor-1alpha. J Biol Chem 277, 21361–21370 (2002).
Cerec, V. et al. Transdifferentiation of hepatocyte-like cells from the human hepatoma HepaRGcell line through bipotent progenitor. Hepatology 45, 957–967 (2007).
Parviz, F. et al. Hepatocyte nuclear factor 4alpha controls the development of a hepatic epithelium and liver morphogenesis. Nat Genet 34, 292–296 (2003).
Battle, M. A. et al. Hepatocyte nuclear factor 4alpha orchestrates expression of cell adhesion proteins during the epithelial transformation of the developing liver. Proc Natl Acad Sci U S A 103, 8419–8424 (2006).
Fahy, E. et al. Update of the LI PID MAPS comprehensive classification system for lipids. J Lipid Res 50 Suppl, S9–S14 (2009).
Subramaniam, S. et al. Bioinformatics and systems biology of the lipidome. Chem Rev 111, 6452–6490 (2011).
Laconi, S. et al. Liver regeneration in response to partial hepatectomy in rats treated with retrorsine: a kinetic study. J Hepatol 31, 1069–1074 (1999).
Samuel, A. & Jago, M. V. Localization in the cell cycle of the antimitotic action of the pyrrolizidine alkaloid, lasiocarpine and of its metabolite, dehydroheliotridine. Chem Biol Interact 10, 185–197 (1975).
Picard, C. et al. Retrorsine: a kinetic study of its influence on rat liver regeneration in the portal branch ligation model. J Hepatol 39, 99–105 (2003).
Rajvanshi, P., Kerr, A., Bhargava, K. K., Burk, R. D. & Gupta, S. Studies of liver repopulation using the dipeptidyl peptidase IV-deficient rat and other rodent recipients: cell size and structure relationships regulate capacity for increased transplanted hepatocyte mass in the liver lobule. Hepatology 23, 482–496 (1996).
Gupta, S. et al. Transplanted hepatocytes proliferate differently after CCl4 treatment and hepatocyte growth factor infusion. Am J Physiol 276, G629–G638 (1999).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chand, L., Risal, P., Shrestha, N. et al. The fate of hepatocyte cell line derived from a liver injury model with long-term in vitro passage. Mol. Cell. Toxicol. 14, 263–272 (2018). https://doi.org/10.1007/s13273-018-0029-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13273-018-0029-x