Abstract
Background
MicroRNAs (miRNAs) could regulate the expression of target genes and play important roles in modulation of various metabolic processes. Nevertheless, little is known about the backfat microRNAome (miRNAome) of the Neijiang pig.
Objectives
The primary objective of this study was to analyse miRNAomes of Landrace and Neijiang pig backfat (LPB and NPB resp.). Furthermore, investigating differentially expressed miRNAs participating in lipid metabolism and mining potential biomarker for Neijiang pig breeding.
Methods
Here we used the Landrace pig with different metabolic characteristics as a control to analyse the Neijiang pig-specific backfat miRNAome. A comprehensive analysis of miRNAomes was performed by deep sequencing.
Results
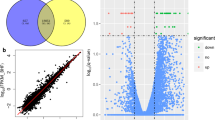
Small RNA sequencing identified 326 unique miRNAs, 280 were co-expressed in both libraries. Only 11 and 35 miRNAs were specifically expressed in LPB and NPB respectively. Sixty seven differentially expressed miRNAs were identified by IDEG6. MiR-1-3p were identified that may participate in lipid metabolism. Furthermore, qPCR results revealed that lower expression of miR-1-3p in NPB could increase the expression of LXRα, which is an enzyme important for the synthesis and accumulation of lipid. The double luciferase report experiment suggested that LXRα was the direct target gene of miR-1-3p. In short, miR-1-3p could modulate the synthesis and accumulation of lipid by target LXRα. It may be a potential marker for pig breeding.
Conclusion
Our investigation has delineated the different miRNAs expression patterns of LPB and NPB, which may help understand the regulatory mechanisms of miRNAs in the lipid metabolism, and provide potential biomarkers for Neijiang pig breeding.






Similar content being viewed by others
References
Ahmad A, Zhang W, Wu M, Tan S, Zhu T (2017) Tumor-suppressive miRNA-135a inhibits breast cancer cell proliferation by targeting ELK1 and ELK3 oncogenes. Genes Genom 40:243–251
Bellingham SA, Coleman BM, Hill AF (2012) Small RNA deep sequencing reveals a distinct miRNA signature released in exosomes from prion-infected neuronal cells. Nucleic Acids Res 21:10937–10949
Bu H, Chen Y, Liu J, Li S, Li Y (2000) RYR1 genotype of the Chinese Neijiang pig. Transpl Proc 32:1058
Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136:642–655
Chan S-P, Slack FJ (2006) Point of View microRNA-mediated silencing inside P-bodies. RNA Biol 3:97–100
Chartoumpekis DV, Zaravinos A, Ziros PG, Iskrenova RP, Psyrogiannis AI, Kyriazopoulou VE, Habeos IG (2012) Differential expression of microRNAs in adipose tissue after long-term high-fat diet-induced obesity in mice. PLoS ONE 7:e34872
Cui S, Cao Z, Guo W, Yu H, Zhou Y (2019) Plasma miRNA-23a and miRNA-451 as candidate biomarkers for early diagnosis of nonsmall cell lung cancer: a case-control study. J South Med Univ 39:705–711
Da Wei Huang BTS, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57
Dávalos A, Goedeke L, Smibert P, Ramírez CM, Warrier NP, Andreo U, Cirera-Salinas D, Rayner K, Suresh U, Pastor-Pareja JC (2011) miR-33a/b contribute to the regulation of fatty acid metabolism and insulin signaling. Proc Natl Acad Sci 108:9232–9237
Enright AJ, John B, Gaul U, Tuschl T, Sander C, Marks DS (2004) MicroRNA targets in Drosophila. Genome Biol 5:R1–R1
Esau C, Davis S, Murray SF, Yu XX, Pandey SK, Pear M, Watts L, Booten SL, Graham M, McKay R (2006) miR-122 regulation of lipid metabolism revealed by in vivo antisense targeting. Cell Metab 3:87–98
Fang Y, Zeng HY, Yun-Cheng LV (2015) MiR-152 inhibits hepatocyte lipid uptake by targeting low density lipoprotein receptor. Chin J Arterioscler 23(8):0774–0778
Fernandez-Valverde SL, Calcino AD, Degnan BM (2015) Deep developmental transcriptome sequencing uncovers numerous new genes and enhances gene annotation in the sponge Amphimedon queenslandica. BMC Genom 16:387
Gang L, Jiansheng W, Sida Q, Xin S, Hong R, Jing Z, Baocheng L (2019) Application of detection of circulating tumor cells and exosome miR-21 in diagnosis of lung ground glass opacity. J Jilin Univ (Med Ed) 299:E198–E206
Gerin I, Bommer GT, McCoin CS, Sousa KM, Krishnan V, MacDougald OA (2010) Roles for miRNA-378/378* in adipocyte gene expression and lipogenesis. Am J Physiol Endocrinol Metab 299:E198–E206
Glazov EA, Cottee PA, Barris WC, Moore RJ, Dalrymple BP, Tizard ML (2008a) A microRNA catalog of the developing chicken embryo identified by a deep sequencing approach. Genome Res 18:957–964
Glazov EA, Kongsuwan K, Assavalapsakul W, Horwood PF, Mitter N, Mahony TJ (2009) Repertoire of bovine miRNA and miRNA-like small regulatory RNAs expressed upon viral infection. PLoS ONE 4:0006349
Gomez-Uchida D, Seeb LW, Warheit KI, Mckinney GJ, Seeb JE (2014) Deep sequencing of the transcriptome and mining of single nucleotide polymorphisms (SNPs) provide genomic resources for applied studies in Chinook salmon (Oncorhynchus tshawytscha). Conserv Genet Resour 6:807–811
Hai B, Min C, Hui C, Du L, Wang Y (2019) Transcriptome-wide identification of miRNA targets and a TAS3-homologous gene in Populus by degradome sequencing. Genes Genom 41:849–861
Huang YP, Guo WZ, Xiao-Qi LI (2007) Molecular cloning, sequencing and constructing eukaryotic expression vector of Neijiang pig IFN-γ. China Anim Husb Vet Med 34:60–63
Iliopoulos D, Drosatos K, Hiyama Y, Goldberg IJ, Zannis VI (2010) MicroRNA-370 controls the expression of MicroRNA-122 and Cpt1α and affects lipid metabolism. J Lipid Res 51:1513–1523
Jingge L, Caibo N, Bojiang L, Rongyang W (2019) Wu H (2018) epatic microRNAome reveals potential microRNA-mRNA pairs association with lipid metabolism in pigs. Asian-Australas J Anim Sci 32(9):1458
Kuchenbauer F, Morin RD, Argiropoulos B, Petriv I, Griffith M, Heuser M, Yung E, Piper J, Delaney A, Prabhu A-L (2008a) In-depth characterization of the microRNA transcriptome in a leukemia progression model. Genome Res 18:1787–1797
Lagos-Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294:853–858
Lai EC (2005) miRNAs: whys and wherefores of miRNA-mediated regulation. Curr Biol 15:R458–R460
Larsen L, Rosenstierne MW, Gaarn LW, Bagge A, Pedersen L, Dahmcke CM, Nielsen JH, Dalgaard LT (2011) Expression and localization of microRNAs in perinatal rat pancreas: role of miR-21 in regulation of cholesterol metabolism. PLoS ONE 6:e25997
Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans. Science 294:858–862
Lehrke M, Lebherz C, Millington SC, Guan HP, Millar J, Rader DJ, Wilson JM, Lazar MA (2005) Diet-dependent cardiovascular lipid metabolism controlled by hepatic LXRα. Cell Metab 1:297–308
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115:787–798
Lian C, Sun B, Niu S, Yang R, Liu B, Lu C, Meng J, Qiu Z, Zhang L, Zhao Z (2012) A comparative profile of the microRNA transcriptome in immature and mature porcine testes using Solexa deep sequencing. FEBS J 279:964–975
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25:402–408
Min F, Wang S, Zhang L (2015) Survey of programs used to detect alternative splicing isoforms from deep sequencing data in silico. Biomed Res Int 2015:1–9
Orangi E, Motovali-Bashi M (2019) Evaluation of miRNA-9 and miRNA-34a as potential biomarkers for diagnosis of breast cancer in Iranian women. Gene 687:272–279
Rehmsmeier M, Steffen P, Höchsmann M, Giegerich R (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10:1507–1517
Romualdi C, Bortoluzzi S, d’Alessi F, Danieli GA (2003) IDEG6: a web tool for detection of differentially expressed genes in multiple tag sampling experiments. Physiol Genomics 12:159–162
Sharbati S, Friedländer MR, Sharbati J, Hoeke L, Chen W, Keller A, Stähler PF, Rajewsky N, Einspanier R (2010) Deciphering the porcine intestinal microRNA transcriptome. BMC Genom 11:275
Takanabe R, Ono K, Abe Y, Takaya T, Horie T, Wada H, Kita T, Satoh N, Shimatsu A, Hasegawa K (2008) Up-regulated expression of microRNA-143 in association with obesity in adipose tissue of mice fed high-fat diet. Biochem Biophys Res Commun 376:728–732
Wang J, Long Y, Zhang J, Xue M, Huang G, Huang K, Yuan Q, Pei X (2018) Combined analysis and miRNA expression profiles of the flowering related genes in common wild rice (oryza rufipogon Griff.). Genes Genom 40(8):835–845
Winter J, Jung S, Keller S, Gregory RI, Diederichs S (2009) Many roads to maturity: microRNA biogenesis pathways and their regulation. Nat Cell Biol 11:228–234
Zhang B, Stellwag EJ, Pan X (2009) Large-scale genome analysis reveals unique features of microRNAs. Gene 443:100–109
Zhao J, Xu J, Zhang R (2018) SRPX2 regulates colon cancer cell metabolism by miR-192/215 via PI3K-Akt. Am J Transl Res 10:483–490
Zhipeng S, Zongde Z, Yang L (2019) Clinical application of plasma miR-34b-3p and miR-302a-5p in the diagnosis of non-small cell lung cancer. Zhongguo fei ai za zhi Chin J Lung Cancer 22(4):216–222
Zhong D, Huang G, Zhang Y, Zeng Y, Xu Z, Zhao Y, He X, He F (2013) MicroRNA-1 and microRNA-206 suppress LXRα-induced lipogenesis in hepatocytes. Cell Signal 25:1429–1437
Acknowledgements
Funding was provided by Doctoral research start up fund of Affiliated Hospital of Southwest Medical University (Grant no. 17138).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that they have no competing interests.
Ethical approval
Animal experiments were performed according to Chinese animal welfare laws and regulations, and approved by the Institutional Animal Care and Use Committee in Neijiang pig breeding farm under Permit No. B2016112.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Li, Y. Comparing of backfat microRNAomes of Landrace and Neijiang pig by high-throughput sequencing. Genes Genom 43, 543–551 (2021). https://doi.org/10.1007/s13258-021-01078-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-021-01078-z