Abstract
Background
Angioimmunoblastic T-cell lymphoma (AITL) is an aggressive disease. Most cancer diagnoses are determined by anatomical histology. Therefore, many samples are stored in FFPE blocks for H&E staining. However, RNAs extracted from the FFPE block have a high level of fragmentation, making it difficult to perform accurate DEG analysis using RNA sequencing.
Objective
To overcome fragmented RNA's drawback in NGS application, we applied the NanoString nCounter® technique of hybridization method that can be used for DEG analysis without PCR amplification.
Methods
We characterized the gene expression profiling of AITLs though transcriptome analysis based on the nCounter® PanCancer IO 360™ Panel and NanoString platform. To perform the analysis of differential expression gene (DEG) profiles in AITLs, we compared the NanoString data from eight AITL patients with a healthy control donor.
Results
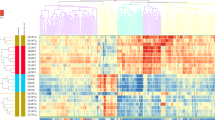
Ninety-one genes were up-regulated and six genes were down-regulated in AITLs compared to control. The Gene Ontology (GO) analysis of 97-DEGs revealed that they were closely related to cytokine, MAPK cascade, leukocyte differentiation, and immune response, suggesting that this affect the immune system. In addition, KEGG analysis revealed that AITL DEGs were found to be highly involved in cytokine–cytokine receptor interaction and PI3K-Akt signaling pathway.
Conclusion
We believe that comprehensive multiplex studies, along with NanoString analysis, may be helpful to understand the molecular mechanisms of AITL, including mutations, gene expression, and protein expression studies.






Similar content being viewed by others
References
Boddicker RL, Razidlo GL, Feldman AL (2019) Genetic alterations affecting GTPases and T-cell receptor signaling in peripheral T-cell lymphomas. Small GTPases 10:33–39
Broccoli A, Zinzani PL (2017) Angioimmunoblastic T-cell lymphoma. Hematol Oncol Clin North Am 31:223–238
Bustelo XR, Sauzeau V, Berenjeno IM (2007) GTP-binding proteins of the Rho/Rac family: regulation, effectors and functions in vivo. BioEssays 29:356–370
Chiba S, Enami T, Ogawa S, Sakata-Yanagimoto M (2015) G17V RHOA: genetic evidence of GTP-unbound RHOA playing a role in tumorigenesis in T cells. Small GTPases 6:100–103
Chihara D, Fanale MA, Miranda RN, Noorani M, Westin JR, Nastoupil LJ, Hagemeister FB, Fayad LE, Romaguera JE, Samaniego F et al (2017) The survival outcome of patients with relapsed/refractory peripheral T-cell lymphoma-not otherwise specified and angioimmunoblastic T-cell lymphoma. Br J Haematol 176:750–758
Cortes JR, Ambesi-Impiombato A, Couronne L, Quinn SA, Kim CS, da Silva Almeida AC, West Z, Belver L, Martin MS, Scourzic L et al (2018) RHOA G17V induces T follicular helper cell specification and promotes lymphomagenesis. Cancer Cell 33(259–273):e257
de Leval L, Rickman DS, Thielen C, Reynies A, Huang YL, Delsol G, Lamant L, Leroy K, Briere J, Molina T et al (2007) The gene expression profile of nodal peripheral T-cell lymphoma demonstrates a molecular link between angioimmunoblastic T-cell lymphoma (AITL) and follicular helper T (TFH) cells. Blood 109:4952–4963
Dobay MP, Lemonnier F, Missiaglia E, Bastard C, Vallois D, Jais JP, Scourzic L, Dupuy A, Fataccioli V, Pujals A et al (2017) Integrative clinicopathological and molecular analyses of angioimmunoblastic T-cell lymphoma and other nodal lymphomas of follicular helper T-cell origin. Haematologica 102:e148–e151
Eastel JM, Lam KW, Lee NL, Lok WY, Tsang AHF, Pei XM, Chan AKC, Cho WCS, Wong SCC (2019) Application of NanoString technologies in companion diagnostic development. Expert Rev Mol Diagn 19:591–598
Etienne-Manneville S, Hall A (2002) Rho GTPases in cell biology. Nature 420:629–635
Evangelista AF, Zanon MF, Carloni AC, de Paula FE, Morini MA, Ferreira-Neto M, Soares IC, Miziara JE, de Marchi P, Scapulatempo-Neto C et al (2017) Detection of ALK fusion transcripts in FFPE lung cancer samples by NanoString technology. BMC Pulm Med 17:86
Fujisawa M, Chiba S, Sakata-Yanagimoto M (2017) Recent progress in the understanding of angioimmunoblastic T-cell lymphoma. J Clin Exp Hematop 57:109–119
Geiss GK, Bumgarner RE, Birditt B, Dahl T, Dowidar N, Dunaway DL, Fell HP, Ferree S, George RD, Grogan T et al (2008) Direct multiplexed measurement of gene expression with color-coded probe pairs. Nat Biotechnol 26:317–325
Han P, Yang L, Yan W, Tian D (2019) Angioimmunoblastic T-cell lymphoma mimicking drug fever and infectious etiology after a thyroidectomy: a case report. Medicine (Baltimore) 98:e16932
Iqbal J, Weisenburger DD, Greiner TC, Vose JM, McKeithan T, Kucuk C, Geng H, Deffenbacher K, Smith L, Dybkaer K et al (2010) Molecular signatures to improve diagnosis in peripheral T-cell lymphoma and prognostication in angioimmunoblastic T-cell lymphoma. Blood 115:1026–1036
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Lemonnier F, Couronne L, Parrens M, Jais JP, Travert M, Lamant L, Tournillac O, Rousset T, Fabiani B, Cairns RA et al (2012) Recurrent TET2 mutations in peripheral T-cell lymphomas correlate with TFH-like features and adverse clinical parameters. Blood 120:1466–1469
Li X, Liu Z, Mi M, Zhang C, Xiao Y, Liu X, Wu G, Zhang L (2019) Identification of hub genes and key pathways associated with angioimmunoblastic T-cell lymphoma using weighted gene co-expression network analysis. Cancer Manag Res 11:5209–5220
Manso R, Roncador G, Montes-Moreno S, Rojo F, Perez-Saenz MA, Mollejo M, Menarguez J, Carvajal N, Garcia-Cosio M, Llamas P et al (2017) p-MAPK1 expression associated with poor prognosis in angioimmunoblastic T-cell lymphoma patients. Br J Haematol 176:661–664
Nakamoto-Matsubara R, Sakata-Yanagimoto M, Enami T, Yoshida K, Yanagimoto S, Shiozawa Y, Nanmoku T, Satomi K, Muto H, Obara N et al (2014) Detection of the G17V RHOA mutation in angioimmunoblastic T-cell lymphoma and related lymphomas using quantitative allele-specific PCR. PLoS One 9:e109714
Ocampo-Garza J, Herz-Ruelas ME, Gonzalez-Lopez EE, Mendoza-Oviedo EE, Garza-Chapa JI, Ocampo-Garza SS, Vazquez-Herrera NE, Miranda-Maldonado I, Ocampo-Candiani J (2014) Angioimmunoblastic T-cell lymphoma: a diagnostic challenge. Case Rep Dermatol 6:291–295
Ohe M, Hashino S (2016) Successful treatment of angioimmunoblastic T-cell lymphoma with clarithromycin. Blood Res 51:139–142
Palomero T, Couronne L, Khiabanian H, Kim MY, Ambesi-Impiombato A, Perez-Garcia A, Carpenter Z, Abate F, Allegretta M, Haydu JE et al (2014) Recurrent mutations in epigenetic regulators, RHOA and FYN kinase in peripheral T cell lymphomas. Nat Genet 46:166–170
Rodriguez-Pinilla SM, Atienza L, Murillo C, Perez-Rodriguez A, Montes-Moreno S, Roncador G, Perez-Seoane C, Dominguez P, Camacho FI, Piris MA (2008) Peripheral T-cell lymphoma with follicular T-cell markers. Am J Surg Pathol 32:1787–1799
Roufosse F, de Leval L, van Krieken H, van Deuren M (2015) Lymphocytic variant hypereosinophilic syndrome progressing to angioimmunoblastic T-cell lymphoma. Leuk Lymph 56:1891–1894
Sakata-Yanagimoto M, Enami T, Yoshida K, Shiraishi Y, Ishii R, Miyake Y, Muto H, Tsuyama N, Sato-Otsubo A, Okuno Y et al (2014) Somatic RHOA mutation in angioimmunoblastic T cell lymphoma. Nat Genet 46:171–175
Schmitz N, de Leval L (2017) How I manage peripheral T-cell lymphoma, not otherwise specified and angioimmunoblastic T-cell lymphoma: current practice and a glimpse into the future. Br J Haematol 176:851–866
Vallois D, Dobay MP, Morin RD, Lemonnier F, Missiaglia E, Juilland M, Iwaszkiewicz J, Fataccioli V, Bisig B, Roberti A et al (2016) Activating mutations in genes related to TCR signaling in angioimmunoblastic and other follicular helper T-cell-derived lymphomas. Blood 128:1490–1502
Veldman-Jones MH, Brant R, Rooney C, Geh C, Emery H, Harbron CG, Wappett M, Sharpe A, Dymond M, Barrett JC et al (2015) Evaluating robustness and sensitivity of the nanostring technologies nCounter platform to enable multiplexed gene expression analysis of clinical samples. Cancer Res 75:2587–2593
Vose J, Armitage J, Weisenburger D, International TCLP (2008) International peripheral T-cell and natural killer/T-cell lymphoma study: pathology findings and clinical outcomes. J Clin Oncol 26:4124–4130
Wang M, Zhang S, Chuang SS, Ashton-Key M, Ochoa E, Bolli N, Vassiliou G, Gao Z, Du MQ (2017) Angioimmunoblastic T cell lymphoma: novel molecular insights by mutation profiling. Oncotarget 8:17763–17770
Watatani Y, Sato Y, Miyoshi H, Sakamoto K, Nishida K, Gion Y, Nagata Y, Shiraishi Y, Chiba K, Tanaka H et al (2019) Molecular heterogeneity in peripheral T-cell lymphoma, not otherwise specified revealed by comprehensive genetic profiling. Leukemia 33:2867–2883
Wickham H (2016) ggplot2: elegant graphics for data analysis. Springer-Verlag, New York
Acknowledgements
The authors gratefully acknowledge Center for Bio-Medical Engineering Core Facility at Dankook University for providing critical reagents and equipment. The present work was conducted with funding from the Research Fund of Dankook University in 2018.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Wonseok Shin, Seyoung Mun, Seungkyu Choi, and Kyudong Han declare that we have no conflict of interest.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13258_2020_919_MOESM1_ESM.pptx
Figure S1. Multidimensional scaling (MDS) plot analysis with the NanoString nCounter gene expression data. The MDS plot analysis was performed to verify similarity of gene expression level between control and AITLs. Blue dot and orange triangle indicate control and ALTL, respectively
13258_2020_919_MOESM2_ESM.pptx
Figure S2. Box plots of the NanoString nCounter gene expression in healthy control and AITLs. The box plots show the distribution of gene expression for each sample, based on the NanoString nCounter data. Blue and orange boxes at same line with x-axis indicate control and ALTL samples, respectively. The Y axis represents a normalized count (transformed to log2). The line in the box represent median and the bars across the boxes show the minimum and maximum expression values in each sample
13258_2020_919_MOESM3_ESM.pptx
Figure S3. GO network analysis of 97 DEGs in AITL. GO network analysis of 97-AITL significant DEGs was performed using the MetaScape web-based tool. Each term is represented by a circle node and its color at right side represents its GO cluster identity. The color intensity of each line reflects statistical significance between the GO clusters
13258_2020_919_MOESM4_ESM.pptx
Figure S4. KEGG network analysis of 97 DEGs in AITL. KEGG network analysis of 97-AITL significant DEGs was performed using the MetaScape web-based tool. Each term is represented by a circle node and its color at right side represents its KEGG cluster identity. The color intensity of each line reflects statistical significance between the KEEG clusters
Rights and permissions
About this article
Cite this article
Shin, W., Mun, S., Choi, S. et al. Application of NanoString technologies in angioimmunoblastic T cell lymphoma. Genes Genom 42, 485–494 (2020). https://doi.org/10.1007/s13258-020-00919-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-020-00919-7