Abstract
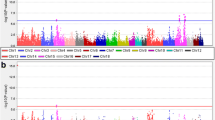
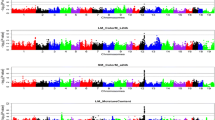
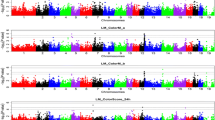
Genome-wide association studies (GWAS) have been steadily used for identification of genomic links to disease and various economical traits. Of those traits, a tenderness of pork is one of the most important factors in quality evaluation of consumers. In this study, we use two pig breed populations; Berkshire is known for its excellent meat quality and Duroc which is known for its high intramuscular fat content in meat. Multivariate genome-wide association studies (MV-GWAS) was executed to compare SNPs of two pigs to find out what genetic variants occur the tenderness of pork. Through MV-GWAS, we have identified candidate genes and the association of biological pathways involved in the tenderness of pork. From these direct and indirect associations, we displayed the usefulness of simple statistical models and their potential contribution to improving the meat quality of pork. We identified a candidate gene related to the tenderness in only Berkshire. Furthermore, several of the biological pathways involved in tenderness in both Berkshire and Duroc were found. The candidate genes identified in this study will be helpful to use them in breeding programs for improving pork quality.


Similar content being viewed by others
References
Andersson L (2001) Genetic dissection of phenotypic diversity in farm animals. Nat Rev Genet 2:130–138
Andersson L, Georges M (2004) Domestic-animal genomics: deciphering the genetics of complex traits. Nat Rev Genet 5:202–212
Davis W, Ohno Y (2010) Color quality scale. Opt Eng 49:033602-033602-033616
De Vries AG, Faucitano L, Sosnicki A, Plastow GS (2000) The use of gene technology for optimal development of pork meat quality. Food Chem 69:397–405
Galesloot TE, van Steen K, Kiemeney LALM., Janss LL, Vermeulen SH (2014) A comparison of multivariate genome-wide association methods. PLoS ONE 9:e95923
Goodwin R, Burroughs S (1995) Genetic evaluation terminal line program results. National Pork Producers Council, Des Moines
Hamill RM, McBryan J, McGee C, Mullen AM, Sweeney T, Talbot A, Cairns MT, Davey GC (2012) Functional analysis of muscle gene expression profiles associated with tenderness and intramuscular fat content in pork. Meat Sci 92:440–450
Iqbal A, Kim YS, Kang JM, Lee YM, Rai R, Jung JH, Oh DY, Nam KC, Lee HK, Kim JJ (2015) Genome-wide association study to identify quantitative trait loci for meat and carcass quality traits in Berkshire. Asian-Australas J Anim Sci 28:1537–1544
Lee T, Shin D-H, Cho S, Kang HS, Kim SH, Lee H-K, Kim H, Seo K-S (2014) Genome-wide association study of integrated meat quality-related traits of the duroc pig breed. Asian-Australas J Anim Sci 27:303–309
Luo W et al (2012) Genome-wide association analysis of meat quality traits in a porcine Large White × Minzhu intercross population. Int J Biol Sci 8:580
Rosenvold K, Andersen HJ (2003) Factors of significance for pork quality—a review. Meat Sci 64:219–237
Van Laack RL, Stevens SG, Stalder KJ (2001) The influence of ultimate pH and intramuscular fat content on pork tenderness and tenderization. J Anim Sci 79:392–397
Wang L, Liu S, Niu T, Xu X (2005) SNPHunter: a bioinformatic software for single nucleotide polymorphism data acquisition and management. BMC Bioinform 6:60
Weisstein EW (2004) Bonferroni correction. Wolfram MathWorld
Weston AR, Rogers RW, Althen TG (2002) The role of collagen in meat tenderness. PAS 18:107–111
Acknowledgements
This work was carried out with the support of “Cooperative Research Program for Agriculture Science & Technology Development (Project No. PJ01111501)” Rural Development Administration, Republic of Korea.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Dongsung Jang declares that he/she does not have conflict of interest on the contents of manuscript. Joon Yoon declares that he/she does not have conflict of interest on the contents of manuscript. Mengistie Taye declares that he/she does not have conflict of interest on the contents of manuscript. Wonseok Lee declares that he/she does not have conflict of interest on the contents of manuscript. Taehyung Kwon declares that he/she does not have conflict of interest on the contents of manuscript. Seunghyun Shim declares that he/she does not have conflict of interest on the contents of manuscript. Heebal Kim declares that he/she does not have conflict of interest on the contents of manuscript.
Ethical approval
The regional Ethical Committees approved whole experiment and all its procedures (JNU Animal Bioethics Committee Approval Number: 2013-0009, National Institute of Animal Science’s Institutional Animal Care and Use Committee: 2009-077).
Rights and permissions
About this article
Cite this article
Jang, D., Yoon, J., Taye, M. et al. Multivariate genome-wide association studies on tenderness of Berkshire and Duroc pig breeds. Genes Genom 40, 701–705 (2018). https://doi.org/10.1007/s13258-018-0672-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-018-0672-6