Abstract
TC1/Mariner transposons belong to class II transposable elements (TEs) that use DNA-mediated “cut and paste” mechanism to transpose, and they have been identified in almost all organisms. Although silkworm (Bombyx mori) has a large amount of TC1/Mariner elements, the genome wide information of this superfamily in the silkworm is unknown. In this study, we have identified 2670 TC1/Mariner (Bmmar) elements in the silkworm genome. All the TEs were classified into 22 families by means of fgclust, a tool of repetitive sequence classification, seven of which was first reported in this study. Phylogenetic and structure analyses based on the catalytic domain (DDxD/E) of transposase sequences indicated that all members of TC1/Mariner were grouped into five subgroups: Mariner, Tc1, maT, DD40D and DD41D/E. Of these five subgroups, maT rather than Mariner possessed most members of TC1/Mariner (51.23%) in the silkworm genome. In particular, phylogenetic analysis and structure analysis revealed that Bmmar15 (DD40D) formed a new basal subgroup of TC1/Mariner element in insects, which was referred to as bmori. Furthermore, we concluded that DD40D appeared to intermediate between mariner and Tc1. Finally, we estimated the insertion time for each copy of TC1/Mariner in the silkworm and found that most of members were dramatically amplified during a period from 0 to 1 mya. Moreover, the detailed functional data analysis showed that Bmmar1, Bmmar6 and Bmmar9 had EST evidence and intact transposases. These implied that TC1/Mariner might have potential transpositional activity. In conclusion, this study provides some new insights into the landscape, origin and evolution of TC1/Mariner in the insect genomes.




Similar content being viewed by others
References
Bryan G, Garza D, Hartl D (1990) Insertion and excision of the transposable element mariner in Drosophila. Genetics 125:103–114
Capy P, Vitalis R, Langin T, Higuet D, Bazin C (1996) Relationships between transposable elements based upon the integrase-transposase domains: is there a common ancestor? J Mol Evol 42:359–368
Capy P, Bazin C, Higuet D, Langin T (1997) Dynamics and evolution of transposable elements. Landes Bioscience, Austin
Clark KJ, Carlson DF, Leaver MJ, Foster LK, Fahrenkrug SC (2009) Passport, a native Tc1 transposon from flatfish, is functionally active in vertebrate cells. Nucleic Acids Res 37:1239–1247
Claudianos C, Brownlie J, Russell R, Oakeshott J, Whyard S (2002) maT:a clade of transposons intermediate between mariner and Tc1. Mol Biol Evol 19:2101–2109
Colloms SD, van Luenen HGAM., PIasterk RHA (1994) DNA binding activities of the Caenorhabditis elegans Tc3 transposase. Nucleic Acids Res 22:5548–5554
Daboussi MJ, Langin T, Brygoo Y (1992) Fot1, a new family of fungal transposable elements. Mol Gen Genet 232:12–16
Delauriere L, Chenais B, Hardivillier Y, Gauvry L, Casse N (2009) Mariner transposons as genetic tools in vertebrate cells. Genetica 137:9–17
Doak TG, Doerder FP, Jahn CL, Herrick G (1994) A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci USA 91:942–946
Duan J, Li RQ, Cheng DJ, Fan W, Zha XF, Cheng TC, Wu YQ, Wang J, Mita K, Xiang ZH, Xia QY (2010) SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res 38:D453-D456
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Emmons SW, Yesner L, Ruan K, Katzenberg D (1983) Evidence for a transposon in Caenorhabditis elegans. Cell 32:55–65
Feschotte C, Wessler SR (2002) Mariner-like transposases are widespread and diverse in flowering plants. Proc Natl Acad Sci USA 99:280–285
Gomulski LM, Torti C, Bonizzoni M, Moralli D, Raimondi E, Capy P, Gasperi G, Malacrida AR (2010) A new basal subfamily of mariner elements in Ceratitis rosa and other tephritid flies. J Mol Evol 53:597–606
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT
Han MJ, Shen YH, Gao YH, Chen LY, Xiang ZH, Zhang Z (2010) Burst expansion, distribution and diversification of MITEs in the silkworm genome. BMC Genom 11:520
Haymer DS, Marsh JL (1986) Germ line and somatic instability of a white mutation in Drosophila mauritiana due to a transposable genetic element. Dev Genet 6:281–291
Jacobson JW, Medhora MM, Hartl DL (1986) Molecular structure of a somatically unstable transposable element in Drosophila. Proc Natl Acad Sci USA 83:8684–8688
Kalendar R, Khassenov B, Ramankulov Y, Samuilova O, Ivanov KI (2017) FastPCR: an in silico tool for fast primer and probe design and advanced sequence analysis. Genomics 109:312–319
Kazazian HH (2004) Mobile elements: drivers of genome evolution. Science 303:1626–1632
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol 16:111–120
Kumaresan G, Mathavan S (2002) RGF-PCR: a technique to isolate different copies of a multi-copy gene. Currentence 82:442–447
Kumaresan G, Mathavan S (2004) Molecular diversity and phylogenetic analysis of mariner-like transposons in the genome of the silkworm Bombyx mori. Insect Mol Biol 13:259–271
Lampe DJ (2010) Bacterial genetic methods to explore the biology of mariner transposons. Genetica 138:499–508
Lohe A, Sullivan D, Hartl D (1996) Genetic evidence for subunit interactions in the transposase of the transposable element mariner. Genetics 144:1087–1095
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16:404–405
Mita K et al (2004) The genome sequence of silkworm, Bombyx mori. DNA Res 11:27–35
Munoz-Lopez M, Siddique A, Bischerour J, Lorite P, Chalmers R, Palomeque T (2008) Transposition of Mboumar-9: identification of a new naturally active mariner-family transposon. J Mol Biol 382:567–572
Novak P, Neumann P, Macas J (2010) Graph-based clustering and characterization of repetitive sequences in next-generation sequencing data. BMC Bioinformatics 11:378
Osanai-Futahashi M, Suetsugu Y, Mita K, Fujiwara H (2008) Genome-wide screening and characterization of transposable elements and their distribution analysis in the silkworm, Bombyx mori. Insect Biochem Mol Biol 38:1046–1057
Pietrokovski S, Henikoff S (1997) A helix-turn-helix DNA-binding motif predicted for transposases of DNA transposons. Mol Gen Genet 254:689–695
Plasterk RHA, Izsvak Z, Ivics Z (1999) Resident aliens: the TC1/Mariner superfamily of transposable elements. Trends Genet 118:527–538
Prasad MD, Nurminsky DL, Nagaraju J (2002) Characterization and molecular phylogenetic analysis of mariner elements from wild and domesticated species of silkmoths. Mol Phylogenet Evol 25:210–217
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16:276–277
Robertson HM (1993) The mariner transposable element is widespread in insects. Nature 362:241–245
Robertson HM, Asplund ML (1996) Bmmarl: a basal lineage of the mariner family of transposable elements in the silkworm moth, Bombyx mori. Insect Biochem Mol Biol 26:945–954
Robertson H, Walden K (2003) Bmmar6, a second mori subfamily mariner transposon from the silkworm moth Bombyx mori. Insect Mol Biol 12:167–171
Robertson HM, Soto-Adames FN, Walden KKO, Avancini RMP, Lampe DJ (1998) The mariner transposons of animals: horizontally jumping genes. Horizontal Gene Transfer 268–284
Shao H, Tu Z (2001) Expanding the diversity of the IS630-Tc1-mariner superfamily: discovery of a unique DD37E transposon and reclassification of the DD37D and DD39D transposons. Genetics 159:1103–1115
Soding J, Biegert A, Lupas AN (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res 33:W244–W248
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
The International Silkworm Genome Consortium (2008) The genome of a lepidopteran model insect, the silkworm Bombyx mori. Insect Biochem Mol Biol 38:1036–1045
Tomita S, Sohn BH, Tamura T (1997) Cloning and characterization of a mariner-like element in the silkworm, Bombyx mori. Genes Genet Syst 72:219–228
Vos JC, Plasterk R (1994) Tc1 transposase of Caenorhabditis elegans is an endonuclease with a bipartite DNA binding domain. EMBO J 13:6125–6132
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, Flavell A, Leroy P, Morgante M, Panaud O et al (2007) A unified classification system for eukaryotic transposable elements. Nat Rev Genet 8:973–982
Xia Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, Zhao P, Zha X, Cheng T, Chai C et al (2004) A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science 306:1937–1940
Zerjal T, Joets J, Alix K, Grandbastien MA, Tenaillon MI (2009) Contrasting evolutionary patterns and target specificities among three Tourist-like MITE families in the maize genome. Plant Mol Biol 71:99–114
Acknowledgements
This work is supported by the National Natural Science Foundation of China (31700318 and 31560308 to ZHH, 31471197 to ZZ).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
Li-Qin Xie declares that she does not have conflict of interest. Ping-Lan Wang declares that e/she does not have conflict of interest. Shen-Hua Jiang declares that he does not have conflict of interest. Ze Zhang declares that he does not have conflict of interest. Hua-Hao Zhang declares that he does not have conflict of interest.
Research involving human and animal rights
This article does not contain any studies with human subjects or animals performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Dataset S1
: Multiple alignments of catalytic domain sequences of each TC1/Mariner family used for phylogenetic analysis. (TXT 7 KB)
Fig. S1
: The outline of the procedure in this study. (PDF 14 KB)
Fig. S2
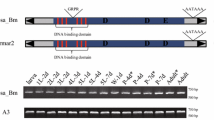
: Structure analysis. As shown that the DNA-binding domain of Bmmars in the silkworm contained six α-helix (black underline), between this two HTH motifs there was a conserved motif GRPR (bright red). DDxD/E domain of these transposases was shown using black letter. DD37D/E, DD40D, and DD41D/E were shown using light green box, deep yellow underline, blue letters, respectively. In this study, we did not consider Bmmar5, Bmmar18, Bmmar19, as transposases of these family that we did not find the DNA binding domain or the catalytic domain. The transposase sequences were aligned by means of MUSCLE program, and then the aligned sequences were modified using the software GeneDoc (http://www.nrbsc.org/gfx/genedoc/whygd.htm) and Illustrator (http://en.wikipedia.org/wiki/Illustrator). (PDF 105 KB)
Table S1
: The information about insertion sites of the TC1/Mariner members into scaffolds. (XLS 401 KB)
Table S2
: TIRs of each TC1/Mariner transposon family. (DOC 73 KB)
Table S3
: Sequence logo of TIRs of TC1/Mariner transposon families created by using WebLogo server. (DOC 1002 KB)
Table S4
: The Accession Number and the matching organisms of each lineage of TC1/Mariner elements. (DOC 67 KB)
Table S5
: The insertion time of each TC1/Mariner copy. (XLS 161 KB)
Table S6
: EST evidence of each TC1/Mariner transposon family. (XLS 21 KB)
Rights and permissions
About this article
Cite this article
Xie, LQ., Wang, PL., Jiang, SH. et al. Genome-wide identification and evolution of TC1/Mariner in the silkworm (Bombyx mori) genome. Genes Genom 40, 485–495 (2018). https://doi.org/10.1007/s13258-018-0648-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-018-0648-6