Abstract
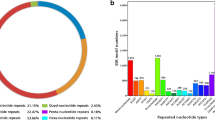
Napier grass (Pennisetum purpureum Schum.) is an economically important species used as a fodder crop, reflecting its high biomass, perennial nature and pest resistance. In order to understand the genomic information and development of this species based on SSR markers, the transcriptome for Napier grass was obtained using next-generation sequencing. A total of 117,076 sequence contigs were obtained, with lengths of 201–10,652 bp. Among these contigs, 44,313 (37.85%) sequences were annotated against public protein databases (E-value <10−5). In addition, the contigs generated from the transcript sequencing were also analysed for the presence of simple sequence repeats (SSRs). A total of 12,744 SSR motifs were identified with in these contigs and corresponding primer pairs were designed. Empirical validation of a cohort of 200 SSRs was performed, with 34% being polymorphic for 20 Napier grass accessions. The polymorphic index content values for each primer ranged from 0.105 to 0.749, with an average of 0.409. The development of genetic, genomic resources and EST-SSRs for Napier grass will contribute to novel gene discovery and the marker-assisted selective breeding of this species.



Similar content being viewed by others
References
Agriculturists N (2009) A healthy aversion to smut. Retrieved Nov 1 98–102
Anderson WF, Dien BS, Brandon SK, Peterson JD (2008) Assessment of bermudagrass and bunch grasses as feedstock for conversion to ethanol. Appl Biochem Biotechnol 145:13–21
Azevedo ALS, Costa PP, Machado JC, Machado MA, Vander Pereira A, da Silva Lédo FJ (2012) Cross species amplification of Pennisetum glaucum microsatellite markers in Pennisetum purpureum and genetic diversity of Napier grass accessions. Crop Sci 52:1776
Babu C, Sundaramoorthi J, Vijayakumar G, Ram SG (2009) Analysis of genetic diversity in napier grass (Pennisetum purpureum Schum) as detected by RAPD and ISSR markers. J Plant Biochem Biotechno 18:181–187
Bayer W (1990) Napier grass–a promising fodder for smallholder livestock production in the tropics. Plant Res Dev 31:103–111
Bhandari AP, Sukanya D, Ramesh C (2006) Application of isozyme data in fingerprinting Napier grass (Pennisetum purpureum Schum.) for germplasm management. Genet Resour Crop Evol 53:253–264
Boa E, Ajanga S, Mulaa M, Jones P (2005) Going Public in Kenya. How to entertain and inform lots of people about napier grass stunt, a new threat to dairy farmers in East Africa. Global Plant Clinic, CABI E-UK, Egham
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32(3):314
Budak H, Shearman R, Parmaksiz I, Dweikat I (2004) Comparative analysis of seeded and vegetative biotype buffalograsses based on phylogenetic relationship using ISSRs, SSRs, RAPDs, and SRAPs. Theor Appl Genet 109:280–288
Burton GW (1989) Registration of ‘Merkeron’napiergrass. Crop Sci 29:1327
Castoe TA, Poole AW, Gu W, Jason de Koning A, Daza JM, Smith EN and Pollock DD (2010) Rapid identification of thousands of copperhead snake (Agkistrodon contortrix) microsatellite loci from modest amounts of 454 shotgun genome sequence. Mol Ecol Resour 10:341–347
Chen D, Li Q, Kong L (2010) Development and characterization of 47 genic microsatellite markers of the loach, Misgurnus anguillicaudatus. J World Aquac Soc 41(1):163–167
Chen H, Wang L, Wang S, Liu C, Blair MW, Cheng X (2015) Transcriptome sequencing of mung bean (Vigna radiate L.) genes and the identification of EST-SSR markers. PLoS ONE 10:e0120273
Coleman TP, Roesser JR (1998) RNA secondary structure: an important cis-element in rat calcitonin/CGRP pre-messenger RNA splicing. Biochem 37(45):15941–15950
de Lima R, Daher R, Goncalves L, Rossi D, do Amaral Júnior A, Pereira M, Lédo F (2011) RAPD and ISSR markers in the evaluation of genetic divergence among accessions of elephant grass. Genet Mol Res 10:1304–1313
Emrich SJ, Barbazuk WB, Li L, Schnable PS (2007) Gene discovery and annotation using LCM-454 transcriptome sequencing. Genome Res 17:69–73
Ferreira ME, Grattapaglia D (1995) Introdução ao uso de marcadores RAPD e RFLP em análise genética. Embrapa-Cenargen, Brasília
Franchini P, Van der Merwe M, Roodt-Wilding R (2011) Transcriptome characterization of the South African abalone Haliotis midae using sequencing-by-synthesis. BMC Res Notes 4:59
Gao Z, Luo W, Liu H, Zeng C, Liu X, Yi S, Wang W (2012a) Transcriptome analysis and SSR/SNP markers information of the blunt snout bream (Megalobrama amblycephala). PLoS ONE 7(8):e42637
Giordano A, Cogan NO, Kaur S, Drayton M, Mouradov A, Panter S, Schrauf GE, Mason JG, Spangenberg GC (2014) Gene discovery and molecular marker development, based on high-throughput transcript sequencing of Paspalum dilatatum Poir. PloS ONE 9:e85050
Hanna WW, Chaparro CJ, Mathews BW, Burns JC, Sollenberger LE, Carpenter JR (2004) Perennial. Warm-season (C4) grasses. American Society of Agronomy, Madison, 503–535
Harris K, Anderson W, Malik R (2010) Genetic relationships among napiergrass (Pennisetum purpureum Schum.) nursery accessions using AFLP markers. Plant Gen Res 8(1):63–70
Huang QL (2008) Studies on the improved variety selection, response to N fertilizers of forages in grass family and their utilization to animal husbandry. Dissertation, University of Fujian Agriculture and Forestry, P.R. China
Jakob K, Zhou F, Paterson AH (2009) Genetic improvement of C4 grasses as cellulosic biofuel feedstocks. In Vitro Cell Dev Biol Plant 45:291–305
Jaradat AA (2010) Genetic resources of energy crops: Biological systems to combat climate change. Aust J Crop Sci 4:309
Jones P, Devonshire B, Holman T, Ajanga S (2004) Napier grass stunt: a new disease associated with a 16SrXI group phytoplasma in Kenya. Plant Pathol 53:519–519
Kandel R, Singh HP, Singh BP, Harris-Shultz KR, Anderson WF (2015) Assessment of genetic diversity in Napier Grass (Pennisetum purpureum Schum.) using microsatellite, single-nucleotide polymorphism and insertion-deletion markers from Pearl Millet (Pennisetum glaucum [L.] R. Br.). Plant Mol Biol Rep 34:265–272
Kantety RV, La Rota M, Matthews DE, Sorrells ME (2002) Data mining for simple sequence repeats in expressed sequence tags from barley, maize, rice, sorghum and wheat. Plant Mol Biol 48:501–510
Kawube G, Alicai T, Otim M, Mukwaya A, Kabirizi J, Talwana H (2014) Resistance of Napier grass clones to Napier grass stunt disease. Afr Crop Sci J 22(3):229–236
Kawube G, Alicai T, Wanjala B, Njahira M, Awalla J, Skilton R (2015) Genetic diversity in Napier Grass (Pennisetum purpureum) assessed by SSR markers. J Agric Sci 7:147
La Rota M, Kantety RV, Yu J-K, Sorrells ME (2005) Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genomics 6:1
Lee M-K, Tsai W-T, Tsai Y-L, Lin S-H (2010) Pyrolysis of napier grass in an induction-heating reactor. J Anal Appl Pyrolysis 88:110–116
Li C, Ling Q, Ge C, Ye Z, Han X (2015) Transcriptome characterization and SSR discovery in large-scale loach Paramisgurnus dabryanus (Cobitidae, Cypriniformes). Gene 557:201–208
Long Y, Wang Y, Wu S, Wang J, Tian X, Pei X (2015) De novo assembly of transcriptome sequencing in Caragana korshinskii Kom. and characterization of EST-SSR markers. PLoS ONE 10:e0115805
Lowe A, Thorpe W, Teale A, Hanson J (2003) Characterisation of germplasm accessions of Napier grass (pennisetum purpureum and P. purpureum × P. glaucum Hybrids) and comparison with farm clones using RAPD. Genet Resour Crop Evol 50:121–132
Metzgar D, Bytof J, Wills C (2000) Selection against frameshift mutations limits microsatellite expansion in coding DNA. Genome Res 10:72–80
Morgante M, Hanafey M, Powell W (2002) Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat Genet 30:194–200
Muldoon DK, Pearson CJ (1979) The hybrid between Pennisetum americanum and Pennisetum purpureum. Herbage Abstracts 49(5):189–199
Mutegi JK, Mugendi DN, Verchot LV, Kung’u JB (2008) Combining napier grass with leguminous shrubs in contour hedgerows controls soil erosion without competing with crops. Agroforest Syst 74:37–49
Nei M, Li W-H (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Parchman TL, Geist KS, Grahnen JA, Benkman CW, Buerkle CA (2010) Transcriptome sequencing in an ecologically important tree species: assembly, annotation, and marker discovery. BMC Genomics 11:1
Peakall R, Gilmore S, Keys W, Morgante M, Rafalski A (1998) Cross-species amplification of soybean (Glycine max) simple sequence repeats (SSRs) within the genus and other legume genera: implications for the transferability of SSRs in plants. Mol Biol Evol 15:1275–1287
Powell W, Machray GC, Provan J (1996) Polymorphism revealed by simple sequence repeats. Trends Plant Sci 1:215–222
Rohlf F (2008) NTSYSpc: numerical taxonomy system, ver. 2.20. Exeter Publishing Ltd, Setauket
Rozen S, Skaletsky H (2000) Primer 3 on the WWW for generalusers and for biologist programmers. In: Krawetz S, Misener S (eds) Bioinformatics methods and protocols: methods in molecular biology. Humana Press, Totowa, New Jersey, pp 365–386
Saha MC, Mian MR, Eujayl I, Zwonitzer JC, Wang L, May GD (2004) Tall fescue EST-SSR markers with transferability across several grass species. Theor Appl Genet 109:783–791
Sanderson M, Reed R, McLaughlin S, Wullschleger S, Conger B, Parrish D, Wolf D, Taliaferro C, Hopkins A, Ocumpaugh W (1996) Switchgrass as a sustainable bioenergy crop. Bioresour Technol 56:83–93
Smith RL, Schweder M, Chowdhury M, Seib J, Schank S (1993) Development and application of RFLP and RAPD DNA markers in genetic improvement of Pennisetum for biomass and forage production. Biomass Bioenerg 5:51–62
Strezov V, Evans TJ, Hayman C (2008) Thermal conversion of elephant grass (Pennisetum Purpureum Schum) to bio-gas, bio-oil and charcoal. Bioresour Technol 99:8394–8399
Struwig M, Mienie C, Van Den Berg J, Mucina L, Buys M (2009) AFLPs are incompatible with RAPD and morphological data in Pennisetum purpureum (Napier grass). Biochem Syst Ecol 37:645–652
Sundaram GR, Babu C et al (2009) Analysis of genetic diversity in napiergrass (Pennisetum purpureum Schum.) as detected by RAPD and ISSR markers. Plant Biochem Biotech 18:181–187
Tangphatsornruang S, Somta P, Uthaipaisanwong P, Chanprasert J, Sangsrakru D, Seehalak W, Sommanas W, Tragoonrung S, Srinives P (2009) Characterization of microsatellites and gene contents from genome shotgun sequences of mungbean (Vigna radiata (L.) Wilczek). BMC Plant Biol 9:1
Thiel T, Michalek W, Varshney R, Graner A (2003) Exploiting EST databases for the development and characterization of gene-derived SSR-markers in barley (Hordeum vulgare L.). Theor Appl Genet 106:411–422
Triwitayakorn K, Chatkulkawin P, Kanjanawattanawong S, Sraphet S, Yoocha T, Sangsrakru D, Chanprasert J, Ngamphiw C, Jomchai N, Therawattanasuk K (2011) Transcriptome sequencing of Hevea brasiliensis for development of microsatellite markers and construction of a genetic linkage map. DNA Res 18(6):471–482
Vera JC, Wheat CW, Fescemyer HW, Frilander MJ, Crawford DL, Hanski I, Marden JH (2008) Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol Ecol 17:1636–1647
Wang B, Zhu P, Yuan Y, Wang C, Yu C, Zhang H, Zhu X, Wang W, Yao C, Zhuang Z (2014) Development of EST-SSR markers related to salt tolerance and their application in genetic diversity and evolution analysis in Gossypium. Genet Mol Res 13:3732–3746
Xie X-M, Zhou F, Zhang X-Q, Zhang J-M (2009) Genetic variability and relationship between MT-1 elephant grass and closely related cultivars assessed by SRAP markers. J genet 88:281–290
Yan X, Zhang X, Lu M, He Y, An H (2015) De novo sequencing analysis of the Rosa roxburghii fruit transcriptome reveals putative ascorbate biosynthetic genes and EST-SSR markers. Gene 561(1):54–62
Zalapa JE, Cuevas H, Zhu H, Steffan S, Senalik D, Zeldin E, McCown B, Harbut R, Simon P (2012) Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am J Bot 99:193–208
Zeng S, Xiao G, Guo J et al (2010) Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et Zucc.) Maxim. BMC Genomics 11(1):1
Zhou F, Dong Z, Xie X (2007) Review on the research and utilization of Pennisetum purpureum cv. Mott in tropical and subtropical areas. Grassl Turf 3:76–82
Acknowledgements
The authors would like to thank Jianxiong Wang for his assistance. This research was financially supported by a grant from the National High Technology Research and Development Program (863 Program, Project No. 2012AA101801). All data have been deposited in the NCBI database (SRA: SRP079814).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human participants
The article does not contain any studies with human participants performed by any of the authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, J., Chen, Z., Jin, S. et al. Development and characterization of simple sequence repeat (SSR) markers based on a full-length cDNA library of Napier Grass (Pennisetum purpureum Schum). Genes Genom 39, 1297–1305 (2017). https://doi.org/10.1007/s13258-017-0536-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13258-017-0536-5