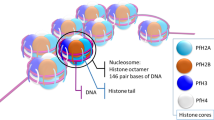
Abstract
Four major human infecting protozoan parasites (Plasmodium, Toxoplasma, Leishmania, and Trypanosoma) impose a substantial threat to health and socio-economic status causing significant morbidity and mortality in tropics and subtropics each year. Lack of effective drugs, the emergence of drug-resistance, and toxicity have made the existing treatment regimen insufficient for most of these parasitic infections ranging from acutely lethal to chronic to almost asymptomatic. These pathogens have developed intricate life-cycle stages altering between multiple hosts and initiating a network of developmental processes in response to environmental stimulus for differentiating within the host without evoking host protective immune surveillance. These differentiation events and successful intracellular survival events require drastic and rapid modulation of the parasites and parasite-driven host gene expression which is achieved by substantial chromatin modifications. Histone posttranslational modifications have a marked effect on chromatin structure organization allowing these pathogens to cope with multiple host survival. In this review, we have described the advancements made in interpreting the role of histone modifications and their impact on gene expression throughout the life-cycle stages of these pathogens. Moreover, we have analyzed available genome-wide transcriptomics datasets for each of these pathogens to identify those parasite-specific histone-modifiers which show a preferential expression in human infective stages, with a perspective of anti-parasitic therapies.



Similar content being viewed by others
Data availability
The data presented in this review will be made available as per journal policy.
Code availability
All the codes used for analyzing genome-wide transcriptomics data are deposited in GitHub 24 (https://github.com/S1403-pi/Histone-modifiers-of-protozoan-parasites.git).
References
Adl SM, Leander BS, Simpson AG, Archibald JM, Anderson OR, Bass D, et al. Diversity, nomenclature, and taxonomy of protists. Syst Biol. 2007;56(4):684–9.
Alcolea PJ, Alonso A, Gomez MJ, Moreno I, Dominguez M, Parro V, et al. Transcriptomics throughout the life cycle of Leishmania infantum: high down-regulation rate in the amastigote stage. Int J Parasitol. 2010;40(13):1497–516.
Allis CD, Jenuwein T. The molecular hallmarks of epigenetic control. Nat Rev Genet. 2016;17(8):487–500.
Alsford S, Horn D. Trypanosomatid histones. Mol Microbiol. 2004;53(2):365–72.
Anderson BA, Wong IL, Baugh L, Ramasamy G, Myler PJ, Beverley SM. Kinetoplastid-specific histone variant functions are conserved in Leishmania major. Mol Biochem Parasitol. 2013;191(2):53–7.
Ataide MA, Andrade WA, Zamboni DS, Wang D, Souza Mdo C, Franklin BS, et al. Malaria-induced NLRP12/NLRP3-dependent caspase-1 activation mediates inflammation and hypersensitivity to bacterial superinfection. PLoS Pathog. 2014;10(1):e1003885.
Belli SI. Chromatin remodelling during the life cycle of trypanosomatids. Int J Parasitol. 2000;30(6):679–87.
Bougdour A, Braun L, Cannella D, Hakimi MA. Chromatin modifications: implications in the regulation of gene expression in Toxoplasma gondii. Cell Microbiol. 2010;12(4):413–23.
Bougdour A, Maubon D, Baldacci P, Ortet P, Bastien O, Bouillon A, et al. Drug inhibition of HDAC3 and epigenetic control of differentiation in Apicomplexa parasites. J Exp Med. 2009;206(4):953–66.
Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem. 2003;278(36):34125–32.
Cavalier-Smith T. Kingdom protozoa and its 18 phyla. Microbiol Rev. 1993;57(4):953–94.
Chaal BK, Gupta AP, Wastuwidyaningtyas BD, Luah YH, Bozdech Z. Histone deacetylases play a major role in the transcriptional regulation of the Plasmodium falciparum life cycle. PLoS Pathog. 2010;6(1):e1000737.
Chandra U, Yadav A, Kumar D, Saha S. Cell cycle stage-specific transcriptional activation of cyclins mediated by HAT2-dependent H4K10 acetylation of promoters in Leishmania donovani. PLoS Pathog. 2017;13(9):e1006615.
Clayton C. Regulation of gene expression in trypanosomatids: living with polycistronic transcription. Open Biol. 2019;9(6):190072.
Cock-Rada AM, Medjkane S, Janski N, Yousfi N, Perichon M, Chaussepied M, et al. SMYD3 promotes cancer invasion by epigenetic upregulation of the metalloproteinase MMP-9. Can Res. 2012;72(3):810–20.
Copeland RA, Solomon ME, Richon VM. Protein methyltransferases as a target class for drug discovery. Nat Rev Drug Discov. 2009;8(9):724–32.
Croken MM, Nardelli SC, Kim K. Chromatin modifications, epigenetics, and how protozoan parasites regulate their lives. Trends Parasitol. 2012;28(5):202–13.
Cui L, Miao J, Furuya T, Li X, Su XZ, Cui L. PfGCN5-mediated histone H3 acetylation plays a key role in gene expression in Plasmodium falciparum. Eukaryot Cell. 2007;6(7):1219–27.
da Cunha JP, Nakayasu ES, de Almeida IC, Schenkman S. Post-translational modifications of Trypanosoma cruzi histone H4. Mol Biochem Parasitol. 2006;150(2):268–77.
de Jesus TC, Nunes VS, Lopes Mde C, Martil DE, Iwai LK, Moretti NS, et al. Chromatin proteomics reveals variable histone modifications during the life cycle of Trypanosoma cruzi. J Proteome Res. 2016;15(6):2039–51.
de Lima LP, Poubel SB, Yuan ZF, Roson JN, Vitorino FNL, Holetz FB, et al. Improvements on the quantitative analysis of Trypanosoma cruzi histone post translational modifications: study of changes in epigenetic marks through the parasite’s metacyclogenesis and life cycle. J Proteomics. 2020;225:103847.
Deshmukh AS, Srivastava S, Dhar SK. Plasmodium falciparum: epigenetic control of var gene regulation and disease. Subcell Biochem. 2013;61:659–82.
DiPaolo C, Kieft R, Cross M, Sabatini R. Regulation of trypanosome DNA glycosylation by a SWI2/SNF2-like protein. Mol Cell. 2005;17(3):441–51.
Divangahi M, Aaby P, Khader SA, Barreiro LB, Bekkering S, Chavakis T, et al. Trained immunity, tolerance, priming and differentiation: distinct immunological processes. Nat Immunol. 2021;22(1):2–6.
Doerig C, Rayner JC, Scherf A, Tobin AB. Post-translational protein modifications in malaria parasites. Nat Rev Microbiol. 2015;13(3):160–72.
Duraisingh MT, Horn D. Epigenetic regulation of virulence gene expression in parasitic protozoa. Cell Host Microbe. 2016;19(5):629–40.
Dzikowski R, Deitsch KW. Genetics of antigenic variation in Plasmodium falciparum. Curr Genet. 2009;55(2):103–10.
Engel JA, Jones AJ, Avery VM, Sumanadasa SD, Ng SS, Fairlie DP, et al. Profiling the anti-protozoal activity of anti-cancer HDAC inhibitors against Plasmodium and Trypanosoma parasites. Int J Parasitol Drugs Drug Resist. 2015;5(3):117–26.
Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15(1):116–27.
Figueiredo LM, Freitas-Junior LH, Bottius E, Olivo-Marin JC, Scherf A. A central role for Plasmodium falciparum subtelomeric regions in spatial positioning and telomere length regulation. EMBO J. 2002;21(4):815–24.
Flueck C, Bartfai R, Volz J, Niederwieser I, Salcedo-Amaya AM, Alako BT, et al. Plasmodium falciparum heterochromatin protein 1 marks genomic loci linked to phenotypic variation of exported virulence factors. PLoS Pathog. 2009;5(9):e1000569.
Fox BA, Guevara RB, Rommereim LM, Falla A, Bellini V, Petre G, et al. Toxoplasma gondii parasitophorous vacuole membrane-associated dense granule proteins orchestrate chronic infection and GRA12 underpins resistance to host gamma interferon. mBio. 2019;10(4).
Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419(6906):498–511.
Gay G, Braun L, Brenier-Pinchart MP, Vollaire J, Josserand V, Bertini RL, et al. Toxoplasma gondii TgIST co-opts host chromatin repressors dampening STAT1-dependent gene regulation and IFN-gamma-mediated host defenses. J Exp Med. 2016;213(9):1779–98.
Guegan F, Bento F, Neves D, Sequeira M, Notredame C, Figueiredo LM. A long non-coding RNA controls parasite differentiation in African trypanosomes. bioRxiv. 2020.
Gupta AP, Zhu L, Tripathi J, Kucharski M, Patra A, Bozdech Z. Histone 4 lysine 8 acetylation regulates proliferation and host-pathogen interaction in Plasmodium falciparum. Epigenetics Chromatin. 2017;10(1):40.
Hollin T, Gupta M, Lenz T, Le Roch KG. Dynamic chromatin structure and epigenetics control the fate of malaria parasites. Trends Genetics TIG. 2021;37(1):73–85.
Horn D. Introducing histone modification in trypanosomes. Trends Parasitol. 2007;23(6):239–42.
Horn D, Cross GA. A developmentally regulated position effect at a telomeric locus in Trypanosoma brucei. Cell. 1995;83(4):555–61.
Horn D, Cross GA. Position-dependent and promoter-specific regulation of gene expression in Trypanosoma brucei. EMBO J. 1997;16(24):7422–31.
Hotez PJ, Alvarado M, Basanez MG, Bolliger I, Bourne R, Boussinesq M, et al. The global burden of disease study 2010: interpretation and implications for the neglected tropical diseases. PLoS Negl Trop Dis. 2014;8(7):e2865.
Hotez PJ, Pecoul B. “Manifesto” for advancing the control and elimination of neglected tropical diseases. PLoS Negl Trop Dis. 2010;4(5):e718.
Jing Q, Cao L, Zhang L, Cheng X, Gilbert N, Dai X, et al. Plasmodium falciparum var gene is activated by its antisense long noncoding RNA. Front Microbiol. 2018;9:3117.
Kaye P, Scott P. Leishmaniasis: complexity at the host-pathogen interface. Nat Rev Microbiol. 2011;9(8):604–15.
Koprinarova M, Botev P, Russev G. Histone deacetylase inhibitor sodium butyrate enhances cellular radiosensitivity by inhibiting both DNA nonhomologous end joining and homologous recombination. DNA Repair. 2011;10(9):970–7.
Kristeleit R, Stimson L, Workman P, Aherne W. Histone modification enzymes: novel targets for cancer drugs. Expert Opin Emerg Drugs. 2004;9(1):135–54.
Kumar D, Saha S. HAT3-mediated acetylation of PCNA precedes PCNA monoubiquitination following exposure to UV radiation in Leishmania donovani. Nucleic Acids Res. 2015;43(11):5423–41.
Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301(5639):1503–8.
Lecoeur H, Prina E, Rosazza T, Kokou K, N’Diaye P, Aulner N, et al. Targeting macrophage histone H3 modification as a leishmania strategy to dampen the NF-kappaB/NLRP3-mediated inflammatory response. Cell Rep. 2020;30(6):1870-82.e4.
Leng J, Denkers EY. Toxoplasma gondii inhibits covalent modification of histone H3 at the IL-10 promoter in infected macrophages. PLoS ONE. 2009;4(10):e7589.
Lerm M, Holm A, Seiron A, Sarndahl E, Magnusson KE, Rasmusson B. Leishmania donovani requires functional Cdc42 and Rac1 to prevent phagosomal maturation. Infect Immun. 2006;74(5):2613–8.
Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5’ flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66(6):1296–305.
Lopez-Rubio JJ, Mancio-Silva L, Scherf A. Genome-wide analysis of heterochromatin associates clonally variant gene regulation with perinuclear repressive centers in malaria parasites. Cell Host Microbe. 2009;5(2):179–90.
Lopez-Rubio JJ, Riviere L, Scherf A. Shared epigenetic mechanisms control virulence factors in protozoan parasites. Curr Opin Microbiol. 2007;10(6):560–8.
Mancio-Silva L, Rojas-Meza AP, Vargas M, Scherf A, Hernandez-Rivas R. Differential association of Orc1 and Sir2 proteins to telomeric domains in Plasmodium falciparum. J Cell Sci. 2008;121(Pt 12):2046–53.
Mandava V, Fernandez JP, Deng H, Janzen CJ, Hake SB, Cross GA. Histone modifications in Trypanosoma brucei. Mol Biochem Parasitol. 2007;156(1):41–50.
Marr AK, MacIsaac JL, Jiang R, Airo AM, Kobor MS, McMaster WR. Leishmania donovani infection causes distinct epigenetic DNA methylation changes in host macrophages. PLoS Pathog. 2014;10(10):e1004419.
Matta SK, Olias P, Huang Z, Wang Q, Park E, Yokoyama WM, et al. Toxoplasma gondii effector TgIST blocks type I interferon signaling to promote infection. Proc Natl Acad Sci USA. 2019;116(35):17480–91.
Miao J, Fan Q, Cui L, Li J, Li J, Cui L. The malaria parasite Plasmodium falciparum histones: organization, expression, and acetylation. Gene. 2006;369:53–65.
Miao J, Fan Q, Cui L, Li X, Wang H, Ning G, et al. The MYST family histone acetyltransferase regulates gene expression and cell cycle in malaria parasite Plasmodium falciparum. Mol Microbiol. 2010;78(4):883–902.
Mukherjee B, Mukhopadhyay R, Bannerjee B, Chowdhury S, Mukherjee S, Naskar K, et al. Antimony-resistant but not antimony-sensitive Leishmania donovani up-regulates host IL-10 to overexpress multidrug-resistant protein 1. Proc Natl Acad Sci USA. 2013;110(7):E575–82.
Naiyer S, Bhattacharya A, Bhattacharya S. Advances in entamoeba histolytica biology through transcriptomic analysis. Front Microbiol. 2019;10:1921.
Nardelli SC, Che FY, Silmon de Monerri NC, Xiao H, Nieves E, Madrid-Aliste C, et al. The histone code of Toxoplasma gondii comprises conserved and unique posttranslational modifications. mBio. 2013;4(6):e00922-13.
Nardelli SC, da Cunha JP, Motta MC, Schenkman S. Distinct acetylation of Trypanosoma cruzi histone H4 during cell cycle, parasite differentiation, and after DNA damage. Chromosoma. 2009;118(4):487–99.
Nast R, Choepak T, Luder CGK. Epigenetic control of IFN-gamma host responses during infection with Toxoplasma gondii. Front Immunol. 2020;11:581241.
Nawaz M, Malik I, Hameed M, Hussain Kuthu Z, Zhou J. Modifications of histones in parasites as drug targets. Vet Parasitol. 2020;278:109029.
Ngwa CJ, Kiesow MJ, Orchard LM, Farrukh A, Llinas M, Pradel G. The G9a histone methyltransferase inhibitor BIX-01294 modulates gene expression during Plasmodium falciparum gametocyte development and transmission. Int J Mol Sci. 2019;20(20).
Nie LB, Liang QL, Elsheikha HM, Du R, Zhu XQ, Li FC. Global profiling of lysine 2-hydroxyisobutyrylome in Toxoplasma gondii using affinity purification mass spectrometry. Parasitol Res. 2020;119(12):4061–71.
Noll TM, Desponds C, Belli SI, Glaser TA, Fasel NJ. Histone H1 expression varies during the Leishmania major life cycle. Mol Biochem Parasitol. 1997;84(2):215–27.
Olias P, Etheridge RD, Zhang Y, Holtzman MJ, Sibley LD. Toxoplasma effector recruits the Mi-2/NuRD complex to repress STAT1 transcription and block IFN-gamma-dependent gene expression. Cell Host Microbe. 2016;20(1):72–82.
Olinski R, Starczak M, Gackowski D. Enigmatic 5-hydroxymethyluracil: oxidatively modified base, epigenetic mark or both? Mutat Res Rev Mutat Res. 2016;767:59–66.
Panneerselvam P, Bawankar P, Kulkarni S, Patankar S. In silico prediction of evolutionarily conserved GC-Rich elements associated with antigenic proteins of Plasmodium falciparum. Evol Bioinform. 2011;7:235–55.
Patil V, Lescault PJ, Lirussi D, Thompson AB, Matrajt M. Disruption of the expression of a non-coding RNA significantly impairs cellular differentiation in Toxoplasma gondii. Int J Mol Sci. 2012;14(1):611–24.
Perez-Toledo K, Rojas-Meza AP, Mancio-Silva L, Hernandez-Cuevas NA, Delgadillo DM, Vargas M, et al. Plasmodium falciparum heterochromatin protein 1 binds to tri-methylated histone 3 lysine 9 and is linked to mutually exclusive expression of var genes. Nucleic Acids Res. 2009;37(8):2596–606.
Petter M, Lee CC, Byrne TJ, Boysen KE, Volz J, Ralph SA, et al. Expression of P. falciparum var genes involves exchange of the histone variant H2A.Z at the promoter. PLoS Pathog. 2011;7(2):e1001292.
Read DF, Cook K, Lu YY, Le Roch KG, Noble WS. Predicting gene expression in the human malaria parasite Plasmodium falciparum using histone modification, nucleosome positioning, and 3D localization features. PLoS Comput Biol. 2019;15(9):e1007329.
Rider SD Jr, Zhu G. Cryptosporidium: genomic and biochemical features. Exp Parasitol. 2010;124(1):2–9.
Robert McMaster W, Morrison CJ, Kobor MS. Epigenetics: a new model for intracellular parasite-host cell regulation. Trends Parasitol. 2016;32(7):515–21.
Roger T, Lugrin J, Le Roy D, Goy G, Mombelli M, Koessler T, et al. Histone deacetylase inhibitors impair innate immune responses to Toll-like receptor agonists and to infection. Blood. 2011;117(4):1205–17.
Roy G, Brar HK, Muthuswami R, Madhubala R. Epigenetic regulation of defense genes by histone deacetylase1 in human cell line-derived macrophages promotes intracellular survival of Leishmania donovani. PLoS Negl Trop Dis. 2020;14(4):e0008167.
Saha S. Histone modifications and other facets of epigenetic regulation in trypanosomatids: leaving their mark. mBio. 2020;11(5).
Saksouk N, Bhatti MM, Kieffer S, Smith AT, Musset K, Garin J, et al. Histone-modifying complexes regulate gene expression pertinent to the differentiation of the protozoan parasite Toxoplasma gondii. Mol Cell Biol. 2005;25(23):10301–14.
Salcedo-Amaya AM, van Driel MA, Alako BT, Trelle MB, van den Elzen AM, Cohen AM, et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc Natl Acad Sci USA. 2009;106(24):9655–60.
Sautel CF, Cannella D, Bastien O, Kieffer S, Aldebert D, Garin J, et al. SET8-mediated methylations of histone H4 lysine 20 mark silent heterochromatic domains in apicomplexan genomes. Mol Cell Biol. 2007;27(16):5711–24.
Scherf A, Lopez-Rubio JJ, Riviere L. Antigenic variation in Plasmodium falciparum. Annu Rev Microbiol. 2008;62:445–70.
Schmidt CQ, Kennedy AT, Tham WH. More than just immune evasion: hijacking complement by Plasmodium falciparum. Mol Immunol. 2015;67(1):71–84.
Schrum JE, Crabtree JN, Dobbs KR, Kiritsy MC, Reed GW, Gazzinelli RT, et al. Cutting edge: Plasmodium falciparum induces trained innate immunity. J Immunol. 2018;200(4):1243–8.
Sindikubwabo F, Ding S, Hussain T, Ortet P, Barakat M, Baumgarten S, et al. Modifications at K31 on the lateral surface of histone H4 contribute to genome structure and expression in apicomplexan parasites. eLife. 2017;6.
Singh U, Ehrenkaufer GM. Recent insights into Entamoeba development: identification of transcriptional networks associated with stage conversion. Int J Parasitol. 2009;39(1):41–7.
Sonda S, Morf L, Bottova I, Baetschmann H, Rehrauer H, Caflisch A, et al. Epigenetic mechanisms regulate stage differentiation in the minimized protozoan Giardia lamblia. Mol Microbiol. 2010;76(1):48–67.
Soto M, Quijada L, Alonso C, Requena JM. Molecular cloning and analysis of expression of the Leishmania infantum histone H4 genes. Mol Biochem Parasitol. 1997;90(2):439–47.
Soto M, Requena JM, Quijada L, Alonso C. Organization, transcription and regulation of the Leishmania infantum histone H3 genes. Biochem J. 1996;318(Pt 3):813–9.
Sullivan WJ Jr, Naguleswaran A, Angel SO. Histones and histone modifications in protozoan parasites. Cell Microbiol. 2006;8(12):1850–61.
Swapna LS, Parkinson J. Genomics of apicomplexan parasites. Crit Rev Biochem Mol Biol. 2017;52(3):254–73.
Tan M, Luo H, Lee S, Jin F, Yang JS, Montellier E, et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell. 2011;146(6):1016–28.
Ukaegbu UE, Kishore SP, Kwiatkowski DL, Pandarinath C, Dahan-Pasternak N, Dzikowski R, et al. Recruitment of PfSET2 by RNA polymerase II to variant antigen encoding loci contributes to antigenic variation in P. falciparum. PLoS Pathog. 2014;10(1):e1003854.
van Luenen HG, Farris C, Jan S, Genest PA, Tripathi P, Velds A, et al. Glucosylated hydroxymethyluracil, DNA base J, prevents transcriptional readthrough in Leishmania. Cell. 2012;150(5):909–21.
Vanagas L, Jeffers V, Bogado SS, Dalmasso MC, Sullivan WJ Jr, Angel SO. Toxoplasma histone acetylation remodelers as novel drug targets. Expert Rev Anti Infect Ther. 2012;10(10):1189–201.
Vanheer LN, Kafsack BFC. Activity comparison of epigenetic modulators against the hemoprotozoan parasites babesia divergens and Plasmodium falciparum. ACS Infect Dis. 2021.
Vembar SS, Scherf A, Siegel TN. Noncoding RNAs as emerging regulators of Plasmodium falciparum virulence gene expression. Curr Opin Microbiol. 2014;20:153–61.
Villares M, Berthelet J, Weitzman JB. The clever strategies used by intracellular parasites to hijack host gene expression. Semin Immunopathol. 2020;42(2):215–26.
Vizuet-de-Rueda JC, Florencio-Martinez LE, Padilla-Mejia NE, Manning-Cela R, Hernandez-Rivas R, Martinez-Calvillo S. Ribosomal RNA genes in the protozoan parasite leishmania major possess a nucleosomal structure. Protist. 2016;167(2):121–35.
Walk J, Keramati F, de Bree LCJ, Arts RJW, Blok B, Netea MG, et al. Controlled human malaria infection induces long-term functional changes in monocytes. Front Mol Biosci. 2020;7:604553.
Wigle TJ. Promoting illiteracy in epigenetics: an emerging therapeutic strategy. Curr Chem Genomics. 2011;5(Suppl 1):48–50.
Yadav A, Chandra U, Saha S. Histone acetyltransferase HAT4 modulates navigation across G2/M and re-entry into G1 in Leishmania donovani. Sci Rep. 2016;6:27510.
Yin D, Jiang N, Zhang Y, Wang D, Sang X, Feng Y, et al. Global lysine crotonylation and 2-hydroxyisobutyrylation in phenotypically different Toxoplasma gondii parasites. Mol Cell Proteomics MCP. 2019;18(11):2207–24.
Yu Z, Genest PA, ter Riet B, Sweeney K, DiPaolo C, Kieft R, et al. The protein that binds to DNA base J in trypanosomatids has features of a thymidine hydroxylase. Nucleic Acids Res. 2007;35(7):2107–15.
Zhang Q, Cao X. Epigenetic regulation of the innate immune response to infection. Nat Rev Immunol. 2019;19(7):417–32.
Zhang Q, Siegel TN, Martins RM, Wang F, Cao J, Gao Q, et al. Exonuclease-mediated degradation of nascent RNA silences genes linked to severe malaria. Nature. 2014;513(7518):431–5.
Acknowledgements
We would like to acknowledge School of Medical Science and Technology, Indian Institute of Technology, Kharagpur for providing all the infrastructure facilities required for this publication.
Funding
SG is a recipient of IIT-KGP GATE fellowship, S is a recipient of CSIR-NET fellowship. BM 14 lab is funded by SERB and MHRD.
Author information
Authors and Affiliations
Contributions
SG participated in preparing the original draft, performed the transcriptome analysis, data preparation, preparing figures, and editing the MS. S participated in preparing the original draft, prepared the figures, and editing the MS. SH performed transcriptome analysis and data preparation. HM participated in preparing and editing the original draft. BM performed supervision, conceptualization, data analyzing, original draft preparation, review, and editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the review was conducted without any conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Corresponding Editor: Somnath Paul; Reviewers: Jonathan B Weitzman, Maumita Bhaumik, Arijit Bhattacharya.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ghosh, S., Snehlata, Hussain, S. et al. Role of chromatin modulation in the establishment of protozoan parasite infection for developing targeted chemotherapeutics. Nucleus 64, 401–413 (2021). https://doi.org/10.1007/s13237-021-00356-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-021-00356-1