Abstract
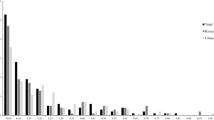
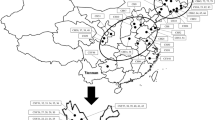
Buckwheat is a pseudocereal that belongs to family Polygonaceae. It is cultivated as a traditional crop in Asia, central Europe and eastern Europe and is used as food as well as medicine. It possesses high quality protein that lacks gluten and is rich in many nutraceuticals and antioxidants; due to which it is presently in great demand. In the present investigation, we collected the buckwheat germplasm from various regions of north western Himalayas of Jammu & Kashmir and Ladakh. The material was purified and then used for molecular variability studies. We used 15 Simple Sequence Repeat (SSRs) to study polymorphism among 52 genotypes of common buckwheat (Fagopyrum esculentum) and further, evaluated these SSRs for transferability in other buckwheat species. Among 15 SSRs isolated from common buckwheat; only seven SSRs could amplify the other buckwheat species. These SSRs were then used to study the genetic variability among 110 genotypes of buckwheat based on the allele frequency. These SSRs amplified a total number of 136 alleles, with 30 alleles amplified by Fem-1322. The major allele frequency ranged from 0.11 to 0.42 and the expected heterozygosity ranged from 0.04 to 0.46. Fem-1322 represents highest Polymorphic Information Content (PIC) of 0.93; followed by Fem-1303 and Fem-1407 with PIC 0.89. The gene diversity ranged from 0.72 to 0.93. Further Darwin based cluster analysis revealed the formation of two major groups by the 110 genotypes. Furthermore, model based STRUCTURE analysis generated two subpopulations which correspond to distance based groups. High level of genetic diversity was observed within the population. The results obtained provided insights about transferability of SSR markers among different species and these findings have further implications in buckwheat breeding as well as conservation strategies.





Similar content being viewed by others
References
Blair MW, Hedetale V, McCouch SR. Fluorescent labeled microsatellite panels useful for detecting allelic diversity in cultivated rice (Oryza sativa L.). Theor Appl Genet. 2009;105:449–57.
Campbell GC. Buckwheat Fagopyrum esculentum Moench. Promoting the conservation and use of underutilized and neglected crops. Institute of Plant Genetics and Crop Plant Research, Gatersleben, Germany and the IPGRI, Rome, Italy; 1997.
Chrungoo NK, Dohtdong L, Chettry U. Genome plasticity in buckwheat. In V. R. Rajpal et al. (eds.). Gene pool diversity and crop improvement. . Sustainable development and biodiversity. Berlin: Springer; 2016, vol. 10, pp. 227–39.
Doyle JJ, Doyle JL. Isolation of plant DNA from fresh tissue. Focus. 1990;12(13):39–41.
Evano G, Regnaut S, Goudet J. Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol. 2005;14:2611–20.
Facho ZH, Farhatullah TW, Ali S. Species divergence and diversity in buckwheat landraces collected from the western Himalayan region of Pakistan. Pak J Bot. 2019;51(6):2215–24.
Goldstein DB, Schlötterer C. Microsatellites. Evolution and application. New York: Oxford University Press; 1999, pp. 449–57.
Hallauer AR, Russell WA, Lamkey KR. Corn breeding. Agronomy Publications; 1988, p. 259.
Ikeda K, Asami Y. Mechanical characteristics of buckwheat noodles. Fagopyrum. 2000;17:67–72.
Iwata H, Imon K, Tsumur Y, Ohsaw R. Genetic diversity among Japanese indigenous common buckwheat (Fagopyrum esculentum) cultivars as determined from amplified fragment length polymorphism and simple sequence repeat markers and quantitative agronomic traits. Genome. 2005;48(3):367–77.
Jing R, Li HQ, Hu CL, Jiang YP, Qin LP, Zheng CJ. Phytochemical and pharmacological profiles of three Fagopyrum buckwheats. Int J Mol Sci. 2016;17(4):589.
Kump B, Javornik B. Evaluation of genetic variability among common buckwheat (Fagopyrum esculentum Moench) populations by RAPD markers. Plant Sci. 1996;114(2):149–58.
Li AR. “Fagopyrum” flora of China. Beijing: Science Press and the Missouri Botanical Garden Press; 2003.
Liu K, Muse SV. Power marker: an integrated analysis environment for genetic marker analysis. Bioinformatics. 2005;21(9):2128–9.
Ma KH, Kim NS, Lee GA, Lee SY, Lee JK, Yi JY, Park YJ, Kim TS, Gwag JG, Kwon SJ. Development of SSR markers for studies of diversity in the genus Fagopyrum. Theor Appl Genet. 2009;119:1247–54.
Marshall HG. Isolation of self-fertile, homomorphic forms in buckwheat, Fagopyrum sagittatum Gilib. Crop Sci. 1969;9:651–3.
Mizuno N, Yasui Y. Gene flow signature in the S-allele region of cultivated buckwheat. BMC Plant Biol. 2019;19:125. https://doi.org/10.1186/s12870-019-1730-1.
de Nettancourt D. Incompatibility and incongruity in wild and cultivated plants. Berlin: Springer; 2001.
Ohnishi O. Search for the wild ancestor of buckwheat III. The wild ancestor of cultivated common buckwheat and of tatary buckwheat. Econ Bot. 1998;52(2):123.
Perrier X, Jacquemoud-Collet JP. DARwin software. https://darwin.cirad.fr/darwin. 2006.
Pritchard JK, Stephens M, Donnelly P. Inference of population structure using multilocus genotype data. Genetics. 2000;155:945–59.
Przybylski R, Gruczyńska E. A review of nutritional and nutraceutical components of buckwheat. Eur J Plant Sci Biotechnol. 2009;3(1):10–22.
Song JY, Lee GA, Yoon MS, Ma KH, Choi YM, Lee JR., Lee MC. Analysis of genetic diversity and population structure of buckwheat (Fagopyrum esculentum Moench.) landraces of Korea using SSR markers. In: Buckwheat germplasm in the world. New York: Academic Press; 2011, pp. 315–31.
Sood M, Chuahan S. Biodiversity conservation in Himalayan region. Int J Appl Res. 2015;2(2):610–5.
Smith JSC, Ertl DS, Orman BA. Identification of maize varieties. In: Wrigley CW, editor. Identification of food grain varieties. St. Paul: American Association for Cereal Chemists; 1995. p. 253–264.
Sytar O, Brestic M, Zivcak M, Phan Tran LS. The contribution of buckwheat genetic resources to health and dietary diversity. Curr Genomics. 2016;17(3):193–206.
Tang Y, Ding MQ, Tang YX, Wu YM, Shao JR, Zhou ML. Germplasm resources of buckwheat in China. In: Zhou M, Kreft I, Woo SH, Chrungoo NK, Wieslander G, editors. Molecular breeding and nutritional aspects of buckwheat. The Netherlands: Academic Press; 2016. p. 13–20.
Tautz D. Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 1989;17(16):6463–71.
Woo SH, Kamal AHM, Tatsuro S, Campbell CG, Adachi T, Yun SH, Chung KY, Choi JS. Buckwheat (Fagopyrum esculentum Moench.): concepts, prospects and potential. Eur J Plant Sci Biotech. 2010;4:1–16.
Wright S. The genetical structure of populations. Ann Eugenics. 1951;15:323–54.
Acknowledgements
SMZ is grateful to NMHS GBPNIHESD, Almora, Uttrakhand, India for financial support of this work (Project Sanction Order No. GBPNI/NMHS17-18/SG24/622).
Author information
Authors and Affiliations
Contributions
EB have contributed in conducting the experiment and preparing first draft of manuscript. RM did the analysis and helped in preparing the manuscript. RAM helped in preparing the manuscript. WAD has helped in collection and field work. SMZ has designed the experiment, helped in analysis, prepared the final manuscript and got grant for undertaking this research work.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Bashir, E., Mahajan, R., Mir, R.A. et al. Unravelling the genetic variability and population structure of buckwheat (Fagopyrum spp.): a collection of north western Himalayas. Nucleus 64, 93–101 (2021). https://doi.org/10.1007/s13237-020-00319-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-020-00319-y