Abstract
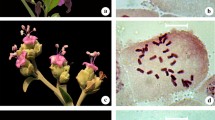
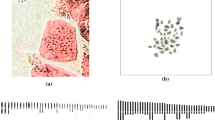
This study was aimed to bring out the Karyotype characteristics of four species of Spilanthes to check for marked symmetry or asymmetry in the chromosomal complements with the aid of semi-automatic image analysis system. The chromosome number of Spilanthes calva and S. radicans was 2n = 78, and in S. ciliata and S. uliginosa it was 2n = 52. The four investigated taxa of Spilanthes are monobasic with the secondary number X 2 = 13. The size of chromosome ranged from 0.50–1.62 μm. The chromosome pairs with secondary constriction are varying from one to three in various taxa. The karyotypes show that considerable variation exists in chromosome morphology and structure among the species studied. In Spilanthes the lesser values for all the parameters of karyomorphology show its rather evolved nature. Such type of variation is important from the evolutionary view point.

Similar content being viewed by others
References
Chung K, Kono Y, Wang CM, Peng CI. Notes on Acmella (Asteraceae: Heliantheae) in Taiwan. Bot Studies. 2008;49:73–82.
Fernandes A, Leitao MT. Contribution a l’etude cytotaxonomique Spermatophyta du Portugal XVII-Lamiaceae. Mem Soc Broteriana. 1984;27:27–75.
Fukui K. Analysis and utility of chromosome information by using chromosome image analyzing system, CHIAS. Bull Natl Inst Agrobiol Resour. 1988;4:153–76.
Fukui K, Kakeda K. Quantitative karyotyping of barley chromosomes by image analysis methods. Genome. 1990;33:450–8.
Fukui K, Kamisugi Y. Mapping of C-banded Crepis chromosomes by imaging methods. Chromosome Res. 1995;3:79–86.
Fukui K, Mukai Y. Condensation pattern as a new image parameter for identification small chromosomes in plants. Jpn J Genet. 1988;63:359–66.
Gottschalk W. Polyploidy and its role in the evolution of higher plants. In: Sharma AK, Sharma A, editor. Advances in chromosome and cell genetics. Oxford: Printsman Press and London: IBH Publ. Co.; 1985.
Grant V. Periodicities in the chromosome numbers of the angiosperms. Bot Gaz. 1982;143:379–89.
Grant V. Plant speciation, 2nd edn. Columbia University Press; 1981. p. 564.
Hind N, Biggs N. Plate 460. Acmella oleracea compositae. Bot Mag. 2003;20:31–9.
Huziwara Y. The karyotype analysis in some genera of Compositae X. The chromosomes of some European species of Aster. Bot Mag. 1962;75:143–50.
Iijima K, Kakeda K, Fukui K. Identification and characterization of somatic rice chromosomes by imaging methods. Theor Appl Genet. 1991;81:591–605.
Jansen RK. Systematic significance of chromosome numbers in Acmella (Asteraceae). Amer J Bot. 1985;72:1835–41.
Jansen RK. The systematics of Acmella (Asteraceae-Heliantheae). Syst Bot Monograph. 1985;8:1–115.
King RM, Robinson H. The Genera of the Eupatorieae (Asteraceae). Monogr Syst Bot Missouri Bot Gard. 1987;22:1–581.
Koopman W, Jong J, De Jong J. A numerical analysis of karyotypes and DNA amounts in Lettuce cultivars and species (Lactuca subsect. Lactuca, Compositae). Acta Bot Neerlandica. 1996;45:211–22.
Krikorian AD, O’connor SA, Fitter MS. Chromosome number variation and Karyotypic stability in cultures and culture derived plants. In: Evans DA, Sharp W R, Ammirato PV, Yamada Y, editors. Hand book of plant cell culture; 1983. p. 541–73.
Lavania UC, Srivastava S. A simple parameter of dispersion index that serves as an adjunct to karyotype asymmetry. J Biosci. 1992;17:179–82.
Levan AK, Fredga K, Sandberg A. Nomenclature for centromeric position on chromosomes. Heriditas. 1964;52:201–20.
Mohanty BD, Ghosh PD, Maity S. Chromosomal analysis in cultured cells of barley (Hordeum Vulgare L.): structural alterations in chromosomes. Cytologia. 1991;56:191–7.
Ojeda MS, Torres LFE. Karyotype analysis of Sporobolus indicus (L.) R. Br. (Ergrostoideae: Gramineae) by image analysis system. Cytologia. 1996;61:301–6.
Rajalakshmi R. Cytological and phytochemical investigations of some medicinal plants of Asteraceae. PhD thesis; 2001
Rajalakshmi R, Joseph J. Karyomorphometrical analysis and Chemical polymorphism in Tagetes erecta and T. patula. Philippine J Sci. 2004;133:135–44.
Sharma AK, Sharma A. Chromosome techniques—theory and practice. 3rd ed. London: Butterworths; 1980.
Watanabe K, Ito M, Yahara T, Sullivan VI, Kawahara T, Crawford DJ. Numerical analyses of Karyotypic diversity in the genes Eupatorium (Composite, Eupatorieae). Plant Syst Evol. 1990;170:215–28.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Rajalakshmi, R., Jose, J. Karyomorphometrical analysis of Spilanthes Jacq. (Asteraceae) using image analysis system. Nucleus 54, 159–168 (2011). https://doi.org/10.1007/s13237-011-0041-1
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13237-011-0041-1