Abstract
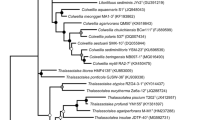
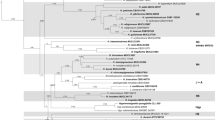
Early efforts to classify Mortierellaceae were based on macro- and micromorphology, but sequencing and phylogenetic studies with ribosomal DNA (rDNA) markers have demonstrated conflicting taxonomic groupings and polyphyletic genera. Although some taxonomic confusion in the family has been clarified, rDNA data alone is unable to resolve higher level phylogenetic relationships within Mortierellaceae. In this study, we applied two parallel approaches to resolve the Mortierellaceae phylogeny: low coverage genome (LCG) sequencing and high-throughput, multiplexed targeted amplicon sequencing to generate sequence data for multi-gene phylogenetics. We then combined our datasets to provide a well-supported genome-based phylogeny having broad sampling depth from the amplicon dataset. Resolving the Mortierellaceae phylogeny into monophyletic genera resulted in 13 genera, 7 of which are newly proposed. Low-coverage genome sequencing proved to be a relatively cost-effective means of generating a high-confidence phylogeny. The multi-gene phylogenetics approach enabled much greater sampling depth and breadth than the LCG approach, but has limitations too. We present this work to resolve some of the taxonomic confusion and provide a genus-level framework to empower future studies on Mortierellaceae diversity and evolution.





Similar content being viewed by others
Data availability
All amplicon sequences and genome sequences have been deposited to Genbank. All trees have been deposited to Treebase (http://purl.org/phylo/treebase/phylows/study/TB2:S25806?x-access-code=16dea5a74941bc1aa812c9ad125aed0&format=html).
Code availability
All custom scripts and pipelines are available on GitHub and/or Zenodo.
References
Ariyawansa HA, Hyde KD, Jayasiri SC, Buyck B, Chethana KT, Dai DQ, Ghobad-Nejhad M et al (2015) Fungal diversity notes 111–252—taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 75(1):27–274
Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, Kulikov AS, Pyshkin AV et al (2012) SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol 19(5):455–477
Beaudet D, Chen EC, Mathieu S, Yildirir G, Ndikumana S, Dalpé Y, Corradi N et al (2017) Ultra-low input transcriptomics reveal the spore functional content and phylogenetic affiliations of poorly studied arbuscular mycorrhizal fungi. DNA Res 25(2):217–227
Benjamin RK (1978) Gamsiella, a new subgenus of Mortierella (Mucorales: Mortierellaceae). Aliso 9(2):157–170
Benny GL (2008) Methods used by Dr. RK Benjamin, and other mycologists, to isolate Zygomycetes. Aliso 26(1):37–61
Benny GL (2009) Zygomycetes. http://www.zygomycetes.org. Accessed 31 Mar 2019
Benny GL, Blackwell M (2004) Lobosporangium, a new name for Echinosporangium Malloch, and Gamsiella, a new genus for Mortierella multidivaricata. Mycologia 96(1):143–149
Blair JE, Coffey MD, Park SY, Geiser DM, Kang S (2008) A multi-locus phylogeny for Phytophthora utilizing markers derived from complete genome sequences. Fungal Genet Biol 45(3):266–277
Bonito G, Hameed K, Ventura R, Krishnan J, Schadt CW, Vilgalys R (2016) Isolating a functionally relevant guild of fungi from the root microbiome of Populus. Fungal Ecol 22:35–42
Brown CT, Irber L (2016) sourmash: a library for MinHash sketching of DNA. J Open Source Softw 1(5):27
Buchfink B, Xie C, Huson DH (2015) Fast and sensitive protein alignment using DIAMOND. Nat Methods 12(1):59
Bushnell B (2014) BBMap. sourceforge.net/projects/bbmap/
Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T (2009) trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25(15):1972–1973
Carreiro MM, Koske RE (1992) Room-temperature isolations can bias against selection of low-temperature microfungi in temperate forest soils. Mycologia 84(6):886–900
Castresana J (2000) Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol 17:540–552 Gblocks version 0.91b
Chang Y, Desirò A, Na H, Sandor L, Lipzen A, Clum A, Smith ME et al (2019) Phylogenomics of Endogonaceae and evolution of mycorrhizas within Mucoromycota. New Phytol 222(1):511–525
Chien CY (1971) A new species of Mortierella from North Carolina. Mycologia 63:826–830
Chien CY (1972) Mortierella umbellata, a new species from Georgia. Mycologia 34:99–102
Chien CY, Kuhlman EG, Gams W (1974) Zygospores in two Mortierella species with ‘Stylospores’. Mycologia 66(1):114–121
de Hoog GS, Guarro J, Gené J, Figueras MJ (2000) Atlas of clinical fungi (edn 2). Centraalbureau voor Schimmelcultures (CBS)
Degawa Y, Tokumasu S (1997) Zygospore formation in Mortierella capitata. Mycoscience 38(4):387–394
Degawa Y, Ohsawa K, Suyama M, Morishita N (2014) Mortierella thereuopodae, a new species with verticillate large sporangiophores, inhabiting fecal pellets of Scutigeromorpha. Mycoscience 55(4):308–313
Detering H, Rutschmann S, Simon S, Fredslund J, Monaghan MT (2016) DiscoMark: Nuclear marker discovery from orthologous sequences using low coverage genome data. bioRxiv, 047282
Dixon-Stewart D (1932) Species of Mortierella isolated from soil. Trans Br Mycol Soc 17:208–220
Domsch KH, Gams W, Anderson TH (1980) Compendium of soil fungi, vol 1. Academic Press, London
Doyle J (1991) DNA protocols for plants. Molecular techniques in taxonomy. Springer, Berlin, pp 283–293
Eddy S (1998) HMMER: profile HMMs for protein sequence analysis
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32(5):1792–1797
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26(19):2460–2461
Finkelstein DB (ed) (2013) Biotechnology of filamentous fungi: technology and products. Newnes, Burlington
Gams W (1976) Some new or noteworthy species of Mortierella. Persoonia 9(1):111–140
Gams W (1977) A key to the species of Mortierella. Persoonia 9(3):381–391
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for basidiomycetes-application to the identification of mycorrhizae and rusts. Mol Ecol 2(2):113–118
GBIF.org (2019) GBIF Home Page. https://www.gbif.org. Accessed 13 Aug 2018
Gerdemann JW, Trappe JM (1974) The Endogonaceae in the Pacific Northwest. The New York Botanical Garden
Goyzueta MLD, Magalhães AI Jr, Ruan Z, de Carvalho JC, Soccol CR (2019) Industrial production, patent landscape, and market trends of arachidonic acid-rich oil of Mortierella alpina. Biotechnol Res Innov. 3:103–109
Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, Wortman JR et al (2008) Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol 9(1):R7
Hibbett D, Glotzer D (2011) Where are all the undocumented fungal species? A study of Mortierella demonstrates the need for sequence-based classification. New Phytol 191(3):592–596
Hopple J, Vilgalys R (1994) Phylogenetic Relationships among Coprinoid Taxa and Allies Based on Data from Restriction Site Mapping of Nuclear rDNA. Mycologia 86(1):96–107. https://doi.org/10.2307/3760723
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ, Lumbsch HT et al (2006) Reconstructing the early evolution of Fungi using a six-gene phylogeny. Nature 443(7113):818
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. https://doi.org/10.1093/nar/gkn201
Jokerst RS, Weeks JR, Zehring WA, Greenleaf AL (1989) Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet MGG 215(2):266–275
Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35:43–46
Kuhlman EG (1969) Mucorales isolated from pine root bark and wood. Can J Bot 47(11):1719–1723
Kuhlman EG (1972) Variation in zygospore formation among species of Mortierella. Mycologia 34:325–341
Kuhlman EG (1975) Zygospore formation in Mortierella alpina and M. spinosa. Mycologia 67(3):678–681
Lanfear R, Frandsen PB, Wright AM, Senfeld T, Calcott B (2016) PartitionFinder 2: new methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol Biol Evol 34(3):772–773
Li H (2018) Minimap2: pairwise alignment for nucleotide sequences. Bioinformatics 34(18):3094–3100
Li GJ, Hyde KD, Zhao RL, Hongsanan S, Abdel-Aziz FA, Abdel-Wahab MA, Baghela A et al (2016) Fungal diversity notes 253–366: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 78(1):1–237
Liao HL, Bonito G, Rojas JA, Hameed K, Wu S, Schadt CW, Vilgalys R et al (2019) Fungal endophytes of Populus trichocarpa alter host phenotype, gene expression, and rhizobiome composition. Mol Plant Microbe Interact 32(7):853–864
Love J, Palmer J, Stajich JE, Esser T, Kastman E, Bogema D, Winter D (2019) nextgenusfs/funannotate: funannotate v1.7.0 (Version 1.7.0). Zenodo. https://doi.org/10.5281/zenodo.3534297
Macias AM, Marek PE, Morrissey EM, Brewer MS, Short DP, Stauder CM, Boyce G (2019) Diversity and function of fungi associated with the fungivorous millipede, Brachycybe lecontii. bioRxiv, 515304
Maddison WP, Maddison DR (2016) Mesquite: a modular system for evolutionary analysis, version 3.10. http://mesquiteproject.org
Malloch D (1967) A new genus of Mucorales. Mycologia 59(2):326–329
Meyer W, Gams W (2003) Delimitation of Umbelopsis (Mucorales, Umbelopsidaceae fam nov) based on ITS sequence and RFLP data. Mycol Res 107:339–350
Miller MA, Pfeiffer W, Schwartz T (2010) Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov 2010, New Orleans, LA, pp 1–8
Mirarab S, Reaz R, Bayzid MS, Zimmermann T, Swenson MS, Warnow T (2014) ASTRAL: genome-scale coalescent-based species tree estimation. Bioinformatics 30(17):i541–i548
Nagy LG, Petkovits T, Kovács GM, Voigt K, Vágvölgyi C, Papp T (2011) Where is the unseen fungal diversity hidden? A study of Mortierella reveals a large contribution of reference collections to the identification of fungal environmental sequences. New Phytol 191(3):789–794
Nampally M, Rajulu MB, Gillet D, Suryanarayanan TS, Moerschbacher BB (2015) A high diversity in chitinolytic and chitosanolytic species and enzymes and their oligomeric products exist in soil with a history of chitin and chitosan exposure. Biomed Res Int. https://doi.org/10.1155/2015/857639
National Center for Biotechnology Information (US), Camacho C (2008) BLAST (r) command line applications user manual. National Center for Biotechnology Information (US)
Olofsson JK, Cantera I, Van de Paer C, Hong-Wa C, Zedane L, Dunning LT, Besnard G et al (2019) Phylogenomics using low-depth whole genome sequencing: a case study with the olive tribe. Mol Ecol Res. https://doi.org/10.1111/1755-0998.13016
Ozimek E, Jaroszuk-Ściseł J, Bohacz J, Korniłłowicz-Kowalska T, Tyśkiewicz R, Słomka A, Hanaka A et al (2018) Synthesis of indoleacetic acid, gibberellic acid and ACC-deaminase by Mortierella strains promote winter wheat seedlings growth under different conditions. Int J Mol Sci 19(10):3218
Palmer, J., & Stajich, J. E. (2017). Funannotate: eukaryotic genome annotation pipeline. Zendo. https://doi.org/10.5281/zenodo.1134477
Petkovits T, Nagy LG, Hoffmann K, Wagner L, Nyilasi I, Griebel T, Papp T et al (2011) Data partitions, Bayesian analysis and phylogeny of the zygomycetous fungal family Mortierellaceae, inferred from nuclear ribosomal DNA sequences. PLoS ONE 6(11):e27507
Pierce NT, Irber L, Reiter T, Brooks P, Brown CT (2019) Large-scale sequence comparisons with sourmash [version 1; peer review: 2 approved]. F1000Research 8:1006. https://doi.org/10.12688/f1000research.19675.1
Price MN, Dehal PS, Arkin AP (2010) FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 5(3):e9490
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19(12):1572–1574
Rudakov OL (1978) Physiological groups in mycophilic fungi. Mycologia 70(1):150–159. https://doi.org/10.1080/00275514.1978.12020210
Seviour RJ, Cooper AL, Skilbeck NW (1987) Identification of Mortierella wolfii, a causative agent of mycotic abortion in cattle. J Med Vet Mycol 25(2):115–123
Shirouzu T, Hirose DD, Tokumasu S (2012) Biodiversity survey of soil-inhabiting Mucoralean and mortierellalean fungi by a baiting method. Jpn J Med Mycol 53:33–39
Sidow A, Thomas WK (1994) A molecular evolutionary framework for eukaryotic model organisms. Curr Biol 4(7):596–603
Slater G (2007) iPCRess. https://github.com/nathanweeks/exonerate
Slater G, Birney E (2005) Automated generation of heuristics for biological sequence comparison. BMC Bioinform 6(1):31
Smith ME, Gryganskyi A, Bonito G, Nouhra E, Moreno-Arroyo B, Benny G (2013) Phylogenetic analysis of the genus Modicella reveals an independent evolutionary origin of sporocarp-forming fungi in the Mortierellales. Fungal Genet Biol 61:61–68
Spatafora JW, Chang Y, Benny GL, Lazarus K, Smith ME, Berbee ML, James TY et al (2016) A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108(5):1028–1046
Stajich, J. E. & Palmer J. M. (2018) AAFTF: Automated Assembly For the Fungi. Zenodo. https://doi.org/10.5281/zenodo.1658103
Stajich, J. E. & Palmer J. M. (2019). Stajichlab/AAFTF: v023 release with (Version v023). Zenodo. http://doi.org/10.5281/zenodo.3437300
Stajich J. E. (2020). 1KFG/PHYling_HMMs_fungi: PHYling markers 13 (Version v13). Zenodo. https://doi.org/10.5281/zenodo.3630031
Stajich JE, Block D, Boulez K, Brenner SE, Chervitz SA, Dagdigian C, Lehväslaiho H et al (2002) The Bioperl toolkit: Perl modules for the life sciences. Genome Res 12(10):1611–1618
Stamatakis A (2014) RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30(9):1312–1313
Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B (2006) AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res 34:435–439
Stockinger H, Peyret-Guzzon M, Koegel S, Bouffaud M-L, Redecker D (2014) The largest subunit of RNA polymerase II as a new marker gene to study assemblages of arbuscular mycorrhizal fungi in the field. PLoS ONE 9(10):e107783. https://doi.org/10.1371/journal.pone.0107783
Takashima Y, Degawa Y, Ohta H, Narisawa K (2018a) Mortierella sugadairana, a new homothallic species related to the firstly described heterothallic species in the genus. Mycoscience 59(3):200–205
Takashima Y, Seto K, Degawa Y, Guo Y, Nishizawa T, Ohta H, Narisawa K (2018b) Prevalence and intra-family phylogenetic divergence of burkholderiaceae-related endobacteria associated with species of Mortierella. Microb Environ 33:417
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30(12):2725–2729
Ter-Hovhannisyan V, Lomsadze A, Chernoff YO, Borodovsky M (2008) Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training. Genome Res 18(12):1979–1990
Thaxter R (1914) New or peculiar Zygomycetes. 3: Blakeslea, Dissophora, and Haplosporangium, nova genera. Bot Gazette 58(4):353–366
Turner M (1956) Mortierella globulifera Rostrup. Trans Br. Mycol Soc 39:291–296
Uehling J, Gryganskyi A, Hameed K, Tschaplinski T, Misztal PK, Wu S, Zienkiewicz K et al (2017) Comparative genomics of Mortierella elongata and its bacterial endosymbiont Mycoavidus cysteinexigens. Environ Microbiol 19(8):2964–2983
Vaidya G, Lohman DJ, Meier R (2011) SequenceMatrix: concatenation software for the fast assembly of multigene datasets with character set and codon information. Cladistics 27(2):171–180
Veerkamp J, Gams W (1983) Los Hongos De Colombia—viii some new species of soil fungi from Colombia. Caldasia 13:709–717
Vilgalys R, Hester M (1990) Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. J Bacteriol 172(8):4238–4246
Wagner L, Stielow B, Hoffmann K, Petkovits T, Papp T, Vágvölgyi C, de Hoog GS, Verkley G, Voigt K (2013) A comprehensive molecular phylogeny of the Mortierellales (Mortierellomycotina) based on nuclear ribosomal DNA. Persoonia 30:77–93
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, Earl AM et al (2014) Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9(11):e112963
Wang L, Chen W, Feng Y, Ren Y, Gu Z, Chen H, Li Y et al (2011) Genome characterization of the oleaginous fungus Mortierella alpina. PLoS ONE 6(12):e28319
Warcup JH (1955) On the origin of colonies of fungi developing on soil dilution plates. Trans Br Mycol Soc 38(3):298–301
Watanabe T (2002) Soil and seed fungi pictorial Atlas of soil and seed fungi. Morphologies of cultured fungi and key to species, 2nd edn. CRC Press, Boca Raton, pp 102–118
Watanabe T, Watanabe Y, Fukatsu T, Kurane R (1998) Mortierella isabellina and M. lignicola from decayed wood with termite nests in Yakushima, Japan. Mycoscience 39(4):475–476
Willoughby LG (1988) Saprolegnia parasitized by Mortierella alpina. Trans Br Mycol Soc 90(3):496–499
Young TWK (1985) Ultrastructure of mucoralean sporangiospores. Bot J Linn Soc 91(1–2):151–165
Zhang F, Ding Y, Zhu CD, Zhou X, Orr MC, Scheu S, Luan YX (2019) Phylogenomics from low-coverage whole-genome sequencing. Methods Ecol Evol 10(4):507–517
Acknowledgements
We extend our gratitude to Amy McGovern and Gail Doehring for assistance in performing DNA extractions, the MGP library preparation and sequencing, and metadata collection. We are grateful to Dr Marty Chilvers, Dr. Andrea Porras-Alfaro, Dr Matthew E Smith, Dr. Matt Kasson and the ZyGoLife project for contributing isolates that were used in these analyses. The collection of Modicella provided by Dr. M.E. Smith was made possible by NSF grants DEB1354802 and DEB1441677. We thank Dr. Kevin Liu for advice on phylogenetic analyses, as well as Bryan Rennick and Alicja Okrasińska for proofreading and helpful discussions.
Disclaimer
The mention of company names or trade products does not imply that they are endorsed or recommended by the US Department of Agriculture over other companies or similar products not mentioned. USDA is an equal opportunity provider and employer.
Funding
US National Science Foundation (NSF) DEB 1737898 (GB and NVP), Michigan State University AgBioResearch NIFA project MICL02416 (GB), NSF STC BEACON Cooperative Agreement DBI-093954 (GB & NVP); US National Science Foundation (The Zygomycetes Genealogy of Life) DEB1354802 and DEB1441677 (JS); JGI-the work conducted by the U.S. Department of Energy Joint Genome Institute, a DOE Office of Science User Facility, is supported by the Office of Science of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231; Data analyses were performed on the High-Performance Computing Cluster at the University of California-Riverside in the Institute of Integrative Genome Biology supported by NSF DBI-1429826 and NIH S10-OD016290 (JS).
Author information
Authors and Affiliations
Contributions
NV-primer design & validation, MLST PCR, MLST sequence analysis, & writing. JL-microscopy, photography, species & genus description, isolate troubleshooting. AD-DNA extraction & strain isolations. HN-LCG sequencing. MK-LCG sequencing. KB-LCG sequencing coordination. IVG-LCG sequencing coordination. ANM-metadata for shared strains & proofreading. KO-idea, strains, research support, MLST sequencing, & proofreading. JES-idea, software development, LCG sequence assembly, annotation, analysis, & writing, data deposition. GB–research ideas, research support, & writing.
Corresponding author
Ethics declarations
Conflict of interest
None to report.
Consent to participate
All authors have agreed to participate in this research
Consent for publication
All authors have read and approved the submitted manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13225_2020_455_MOESM1_ESM.xlsx
Supplementary Table 1: Species not included. A summary of the described species not included in this study, the estimated placement under the proposed taxonomy, the basis for the estimation, and the reference for the original species definition. Supplementary material 1 (XLSX 79 kb)
13225_2020_455_MOESM2_ESM.xlsx
Supplementary Table 2: Isolate metadata. The substrate, geographic origin, collector, collection year, vouchered by, and synonymous isolate identification numbers known for each isolate used in this study. Supplementary material 2 (XLSX 93 kb)
13225_2020_455_MOESM3_ESM.xlsx
Supplementary Table 3: LCG assembly & annotation. The LCG assembly statistics, BUSCO analysis on the fungi_odb9 dataset which contained 290 single-copy marker genes, protein ortholog detection, number of markers used out of 434 total, and assembly deposition/accession details. Sample identifications were adjusted to correct misidentified samples or outdated taxonomy. “REF” indicates reference de novo genomes that were used to guide LCG sequence analysis, which are best accessed by BioSample number. Supplementary material 3 (XLSX 101 kb)
13225_2020_455_MOESM4_ESM.xlsx
Supplementary Table 4: Primer sets. All primer sets produced by the MGP locus selection pipeline. Status indicates which tests the primer set has passed: in silico = simulated PCR in IPCRESS; in vitro = amplification & sequencing of each independent primer set using genomic DNA from a panel of isolates; in vivo = multiplex PCR and sequencing to generate mutli-gene phylogenetic data. Failure at each stage came in the form of 1) non-specific in silico “amplification”, 2) off target in vitro amplification or failure to amplify across the panel of isolates, and 3) in vivo MGP sequence data analysis revealing selective pressure or potential gene duplication of that locus. Supplementary material 4 (XLSX 21 kb)
13225_2020_455_MOESM5_ESM.xlsx
Supplementary Table 5: Raw sequences per locus. The total number of sequences recovered, the total number of isolates represented, and the ratio between the number of sequences and isolates, full and partial length, for each locus. Supplementary material 5 (XLSX 83 kb)
13225_2020_455_MOESM6_ESM.xlsx
Supplementary Table 6: Rejected Strains. The strain number, preliminary identification, ITS-based identification, number of full-length sequences for each locus, and reason the strain was excluded from the final MGP analyses. Supplementary material 6 (XLSX 75 kb)
13225_2020_455_MOESM7_ESM.xlsx
Supplementary Table 7: MGP loci by isolate. Values indicate the Genbank reference number for the sequence included in the final dataset. Numbers in parentheses indicate the initial degree of sequence duplication for that sample at that locus. “0.5” indicates at least one partial sequence was detected but could not be included due to insufficient length. Asterisks indicate sequences obtained from a low coverage or de novo genome sequence, rather than PCR amplification. Supplementary material 7 (XLSX 77 kb)
13225_2020_455_MOESM8_ESM.xlsx
Supplementary Table 8: Primer mismatch. The total number of isolates belonging to each of the ITS-based clades defined by Wagner et al. (2013), the number of those detected in each locus, and the number of usable isolates included in the phylogenetic analyses. The larger the discrepancy between total and detected isolates, the poorer the performance of the primer set on this lineage of Mortierellomycotina. Supplementary material 8 (XLSX 75 kb)
13225_2020_455_MOESM9_ESM.xlsx
Supplementary Table 9: Locus Sequence Variability at the Species Level. Summary statistics from pairwise blastn analyses of the locus sequences as submitted to Genbank. Empty cells indicate that the locus was not recovered for any strains in that species. ‘n’ = the number of non-self pairwise blastn analyses conducted, where ‘0’ indicates that only one sequence was available to represent the species and therefore no intraspecific analyses could be conducted. “Min % - Max %” = the range of percent sequence variability, where ‘0’ means the sequences were identical. Values in bold indicate a meaningful difference between the maximum observed intraspecific variation and the minimum observed interspecific variation. Supplementary material 9 (XLSX 20 kb)
13225_2020_455_MOESM10_ESM.xlsx
Supplementary Table 10: Locus Sequence Variability at the Genus Level. Summary statistics from pairwise blastn analyses of the locus sequences as submitted to Genbank. Empty cells indicate that the locus was not recovered for any strains of any species in that genus. ‘n’ = the number of non-self pairwise blastn analyses conducted, where ‘0’ indicates that only one sequence was available to represent the genus and therefore no intrageneric analyses could be conducted. “Min % - Max %” = the range of percent sequence variability, where ‘0’ means the sequences were identical. Values in bold indicate a meaningful difference between the maximum observed intrageneric variation and the minimum observed intergeneric variation. Supplementary material 10 (XLSX 14 kb)
13225_2020_455_MOESM11_ESM.xlsx
Supplementary Table 11: Species characteristics. The current and proposed classification, synonyms, geographic distribution, ecology, and spore morphologies of Mortierellaceae species represented in this study. Supplementary material 11 (XLSX 82 kb)
13225_2020_455_MOESM12_ESM.xlsx
Supplementary Table 12: Comparative phylogenies. A comparison of the Mortierellaceae phylogenies generated based on the species included in this study, Wagner et al. (2013), Petkovits et al. (2011), and the taxonomic groupings of Linnemann and Gams published in 1977–1989. Supplementary material 12 (XLSX 81 kb)
13225_2020_455_MOESM13_ESM.tif
Supplementary Fig. 1: Unconstrained MGP Mortierellaceae phylogeny. Unconstrained Maximum Likelihood analysis of the concatenated 6-gene MGP dataset (8181 characters). Taxa are named according to the initial ITS-based species identification and current taxonomy. Clade colors indicate monophyletic groupings according to the proposed taxonomy, lines and names denote previously defined clades for the purpose of discussion. Supplementary material 13 (TIFF 113994 kb)
13225_2020_455_MOESM14_ESM.tif
Supplementary Fig. 2: MrBayes MGP Mortierellaceae phylogeny. A Bayesian analysis of the concatenated MGP dataset using a series of partial constraints defined by major nodes in the LCG phylogeny. Clade colors indicate groupings according to the Constrained RAxML MGP phylogeny. Supplementary material 14 (TIFF 413623 kb)
Rights and permissions
About this article
Cite this article
Vandepol, N., Liber, J., Desirò, A. et al. Resolving the Mortierellaceae phylogeny through synthesis of multi-gene phylogenetics and phylogenomics. Fungal Diversity 104, 267–289 (2020). https://doi.org/10.1007/s13225-020-00455-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-020-00455-5