Abstract
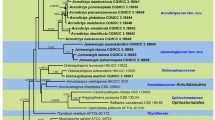
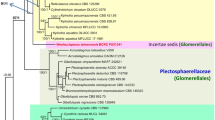
Chalara is a genus of anamorphic fungi with typical phialidic conidiogenous cells. Species of Chalara live mostly as litter saprotrophs, many of them on coniferous litter. In this study, the morphology and DNA sequences (ITS rDNA, 28S rDNA and EF-1α) of strains of various species of Chalara isolated from coniferous litter needles were compared with those of strains from public collections. The majority of the isolates belonged to the C. longipes. Other strains represent C. piceae-abietis, C. microspora, C. microchona and four hitherto undescribed species. These are introduced here as C. holubovae sp. nov., C. hyalocuspica sp. nov., C. pseudoaffinis sp. nov. and C. recta sp. nov. Chalara recta is most closely related to C. longipes, which was found to be paraphyletic. However, no correlation of molecular data with the morphology was found. Chalara holubovae is specific among the Chalara species in that it has a synanamorph with fusiform conidia. Together with C. hyalocuspica, C. holubovae likely belong to the anamorphic Hyaloscyphaceae. Chalara piceae-abietis is epitypified. Epitypicifation of C. longipes and C. microspora cannot be done due to absence of a convenient specimen. Chalara austriaca may be re-discovered also after a targeted sampling in the locality of the type specimen. The majority of studied species are saprotrophic and colonise litter needles. An endophytic phase in living needles or other parts of a tree was confirmed for the species C. longipes and C. hyalocuspica. C. holubovae has been recorded only as an endophyte.









Similar content being viewed by others
References
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Anonymus (1964) ISCC-NBS color-name charts illustrated with centroid colors. The NBS/IBCC Color System—http://www.anthus.com/Colors.html. Accessed 10 July 2010
Aras S, Cansaran D (2006) Isolation of DNA for sequence analysis from herbarium material of some lichen specimens. Turk J Bot 30:449–453
Bogale M, Orr M-J, O’Hara MJ, Untereiner W (2010) Systematics of Catenulifera (anamorphic Hyaloscyphaceae) with an assessment of the phylogenetic position of Phialophora hyalina. Fun Biol 114:396–409
Brock PM, Döring H, Bidartondo MI (2009) How to know unknown fungi: the role of a herbarium. New Phytol 181:719–724
Cai L, Wu W-P, Hyde KD (2009) Phylogenetic relationships of Chalara and allied species inferred from ribosomal DNA sequences. Mycol Prog 8:133–143
Coetsee C, Wingfield MJ, CrousPW WBD (2000) Xenochalara, a new genus of dematiaceous hyphomycetes for chalara-like fungi with apical wall building conidial development. S Afr J Bot 66:99–103
Cole GT, Kendrick WB (1973) Taxonomic studies of Phialophora. Mycologia 3:661–688
Corda ACI (1838) Icones fungorum hucusque cognitorum. Tomus II
Cubero OF, Crespo A, Fatehi J, Bridge PD (1999) DNA extraction and PCR amplification method suitable for fresh, herbarium-stored, lichenized, and other fungi. P1 Syst Evol 216:243–249
de Hoog GS, Vicente V, Caligiorne RB, Kantarcioglu S, Tintelnot K, Gerrits van den Ende AHG, Haase G (2003) Species diversity and polymorphism in the Exophiala spinifera clade containing opportunistic black yeast-like fungi. J Clin Microbiol 41:4767–4778
Edgar RC (2004) MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinform 5:113
Etayo J, Sancho LG (2008) Hongos liquenícolas del Sur de Sudamérica, especialmente de Isla Navarino (Chile). Bibl Lichenol 98:1–302
Fassatiová O (1986) Moulds and filamentous fungi in technical microbiology. Elsevier, Amsterdam
Fautrey MF, Lambotte M (1895) Espèces ou forms nouvelles de la Côte-d’Or. Rev Mycol 17:69–82
Gams W, Holubová-Jechová V (1976) Chloridium and some other dematiaceous hyphomycetes growing on decaying wood. Stud Mycol 13:1–99
Gams W, Philippi S (1992) A study of Cyathicula strobilina and its Chalara anamorph in vitro. Persoonia 14:547–552
Ganley RJ, Newcombe G (2006) Fungal endophytes in seeds and needles of Pinus monticola. Mycol Res 110:318–327
Góes-Neto A, Loguercio-Leite C, Guerrero RT (2005) DNA extraction from frozen field-collected and dehydrated herbarium fungal basidiomata: performance of SDS and CTAB-based methods. Biotemas 18:19–32
Gremmen J (1959) A contribution to the Mycoflora of the pine forests in the Netherlands. N Hedwig 1:251–284
Guindon S, Gascuel O (2003) A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol 52:696–704
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Holubová-Jechová V (1984) Lignicolous Hyphomycetes from Czechoslovakia 7. Chalara, Exochalara, Fusichalara and Dictyochaeta. Fol Geob Phytotaxon 19:387–438
Kile GA, Walker J (1987) Chalara australis sp. nov. (Hyphomycetes), a vascular pathogen of Nothofagus cunninghamii (Fagaceae) in Australia and its relationship to other Chalara species. Aust J Bot 35:1–32
Kirk PM (1986) New or interesting microfungi. XV. Miscellaneous hyphomycetes from the British isles. Trans Br Mycol Soc 86:409–428
Koukol O (2010) Revision of “Septonema ochraceum” revealed three new species of Venturiaceae and Herpotrichiellaceae. Mycol Progress 9:369–378
Kowalski T (2006) Chalara fraxinea sp. nov. associated with dieback of ash (Fraxinus excelsior) in Poland. For Pathol 36:264–270
Kowalski T, Holdenrieder O (2009) The teleomorph of Chalara fraxinea, the causal agent of ash dieback. For Pathol 39:304–308
Kumar S, Tamura K, Nei M (2004) MEGA3: integrated software for molecular evolutionary genetics analysis molecular evolutionary genetics analysis and sequence alignment. Brief Bioinform 5:150–163
Lygis V, Vasiliauskas R, Stenlid J, Vasiliauskas A (2004) Silvicultural and pathological evaluation of Scots pine afforestations mixed with deciduous trees to reduce the infections by Heterobasidion annosum s.s. For Ecol Manag 201:275–285
McKenzie EHC, Pinnoi A, Wong MKM, Hyde KD, Gareth Jones EB (2002) Two new hyaline Chalara species, and a key to species described since 1975. Fun Div 11:129–139, 35:90–101
Minter DW (1981) Microfungi on needles, twigs and cones of pines in ČSSR. Čes Mykol 35:90–101
Nag Raj TR, Kendrick WB (1971) On the identity of three species of Cylindrosympodium described by Preuss. Can J Bot 49:2119–2122
Nag Raj TR, Kendrick WB (1975) A monograph of Chalara and allied genera. Wilfrid Laurier University Press, Waterloo
Nag Raj TR, Kendrick WB (1993) The anamorph as generic determinant in the holomorph: the Chalara connection in the ascomycetes, with special reference to the Ophiostomatoid fungi. In: Wingfield MJ, Seifert KA, Webber JF (eds) Ceratocystis and Ophiostoma: taxonomy, ecology and pathology. St Paul, Minnesota, pp 71–74
Nikolcheva LG, Bärlocher F (2004) Taxon-specific fungal primers reveal unexpectedly high diversity during leaf decomposition in a stream. Mycol Prog 3:41–49
O’Donnell K (1993) Fusarium and its near relatives. In: Reynolds DR, Taylor JW (eds) The fungal holomorph: mitotic, meiotis and pleomorphic speciation in fungal systematics. CAB International, Wallingford, pp 225–233
O’Gorman DT, Sholberg PL, Stokes SC, Ginns J (2008) DNA sequence analysis of herbarium specimens facilitates the revival of Botrytis mali, a postharvest pathogen of apple. Mycologia 100:227–235
Paulin AE, Harrington TC (2000) Phylogenetic placement of anamorphic species of Chalara among Ceratocystis species and other ascomycetes. Stud Mycol 45:209–222
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 49:817–818
Preuss GT (1851) Uebersicht untersuchter Pilze, besonders aus der Umgegend von Hoyerswerda. Linnaea 24:99–153
Punja ZK, Sun LJ (1999) Morphological and molecular characterization of Chalara elegans (Thielaviopsis basicola) cause of black root rot on diverse plant species. Can J Bot 77:1801–1812
Raja HA, Miller AN, Shearer CA (2008) Freshwater ascomycetes: Aquapoterium pinicola, a new genus and species of Helotiales (Leotiomycetes) from Florida. Mycologia 100:141–148
Réblová M (1999) Teleomorph-anamorph connections in Ascomycetes 2. Ascochalara gabretae gen. et sp. nov. and its Chalara-like anamorph. Sydowia 51:210–222
Rehner SA, Buckley E (2005) A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97:84–98
Ronquist F, Huelsenbeck JP (2003) MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19:1572–1574
Telle S, Thines M (2008) Amplification of cox2 (∼620 bp) from 2 mg of up to 129 years old herbarium specimens, comparing 19 extraction methods and 15 polymerases. PLoS ONE 3(10):e3584. doi:10.1371/journal.pone.0003584
Wang Z, Binder M, Schoch CL, Johnston PR, Spatafora JW, Hibbett DS (2006) Evolution of helotialean fungi (Leotiomycetes, Pezizomycotina): a nuclear rDNA phylogeny. Mol Phylogenet Evol 41:295–312
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic, San Diego, pp 315–322
Wu W-P (2004) Chalara and allied genera from China: new species and new records of Chalara from China. Mycosystema 23:313–323
Acknowledgements
I am very grateful to Dr. Miroslav Kolařík for participation in sequencing and valuable comments on the molecular analyses, Zuzana Kolářová for isolation of Chalara from spruce litter and to Dr. Roland Kirschner for sending two specimens of Chalara. This study was supported by the Grant Agency of the Czech Republic, project No. 206/09/P295 and 526/08/0751.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Koukol, O. New species of Chalara occupying coniferous needles. Fungal Diversity 49, 75–91 (2011). https://doi.org/10.1007/s13225-011-0092-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-011-0092-2