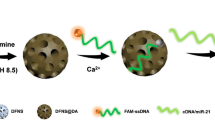
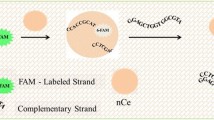
Abstract
Carbon nanotubes (CNTs) are versatile materials that act as natural fluorescence quenchers and double scaffolds for DNA that can wrap around them based on π–π stacking. We exploited these properties of CNTs to develop a hybridization-based sensor for the detection of microRNA. We designed a fluorescein amidite (FAM)-labeled single-stranded oligonucleotide containing a CNT binding region (Poly T) followed by a sequence complementary to the target nucleic acid (probe sequence). As the DNA wraps around a CNT, FAM fluorescence is quenched in the absence of the target, whereas in the presence of the target, fluorescence emission is obtained. Experimentally, we found that one of the major issues with this sensor is its compromised sensitivity due to competition for adsorption of the probe DNA onto the CNT versus that of hybridization with the target DNA. To overcome this, we introduced a short complementary sequence (SCS) that binds to the probe sequence and found that it significantly improved the limit of detection of the sensor approximately 25-fold. To gain further insights into the mechanism of SCS in improving the sensor performance, we performed molecular dynamics (MD)-based simulations. Based on hybridization energy calculations performed using MM-GBSA, we found that the position of the SCS is key to shaping the binding affinity of the probe to the CNT. The MD-based calculations were validated using experimental results by comparing the sensor’s experimental limit of detection and the hybridization energy obtained computationally. Finally, we demonstrated the applicability of this sensor for the experimental detection of the cervical cancer-related biomarker miR-21-5p in human serum.






Similar content being viewed by others
References
Kim, J., Park, M.: Recent progress in electrochemical immunosensors. Biosensors 11, 360 (2021)
Park, M., Heo, Y.J.: Biosensing technologies for chronic diseases. BioChip J. 15, 1–13 (2021)
Cho, S.-Y., Jin, X., Gong, X., Yang, S., Cui, J., Strano, M.S.: Antibody-free rapid detection of SARS-CoV-2 proteins using corona phase molecular recognition to accelerate development time. Anal. Chem. 93, 14685–14693 (2021)
Ferrier, D.C., Honeychurch, K.C.: Carbon nanotube (CNT)-based biosensors. Biosensors 11, 486 (2021)
Lee, W.S., Ahn, J., Jung, S., Lee, J., Kang, T., Jeong, J.: Biomimetic nanopillar-based biosensor for label-free detection of Influenza A virus. BioChip J. 15, 260–267 (2021)
Baek, J.M., Ryu, Y.-S.: Surface sensitive analysis device using model membrane and challenges for biosensor-chip. BioChip J. 14, 110–123 (2020)
Li, C., Shi, G.: Carbon nanotube-based fluorescence sensors. J. Photochem. Photobiol. C: Photochem. Rev. 19, 20–34 (2014)
Kruss, S., Hilmer, A.J., Zhang, J., Reuel, N.F., Mu, B., Strano, M.S.: Carbon nanotubes as optical biomedical sensors. Adv. Drug Deliv. Rev. 65, 1933–1950 (2013)
Kim, J., Hwang, E.-S.: Multiplexed diagnosis of four serotypes of dengue virus by real-time RT-PCR. BioChip J. 14, 421–428 (2020)
Bachilo, S.M., Strano, M.S., Kittrell, C., Hauge, R.H., Smalley, R.E., Weisman, R.B.: Structure-assigned optical spectra of single-walled carbon nanotubes. Science 298, 2361–2366 (2002)
Zhu, Z., Yang, R., You, M., Zhang, X., Wu, Y., Tan, W.: Single-walled carbon nanotube as an effective quencher. Anal. Bioanal. Chem. 396, 73–83 (2010)
Swathi, R.S., Sebastian, K.L.: Resonance energy transfer from a dye molecule to graphene. J. Chem. Phys. 129, 054703 (2008)
Murakami, H., Nomura, T., Nakashima, N.: Noncovalent porphyrin-functionalized single-walled carbon nanotubes in solution and the formation of porphyrin-nanotube nanocomposites. Chem. Phys. Lett. 378, 481–485 (2003)
Fowler, P., Ceulemans, A.: Electron deficiency of the fullerenes. J. Phys. Chem. 99, 508–510 (1995)
Zheng, M., Jagota, A., Strano, M.S., Santos, A.P., Barone, P., Chou, S.G., Diner, B.A., Dresselhaus, M.S., McLean, R.S., Onoa, G.B., Samsonidze, G.G., Semke, E.D., Usrey, M., Walls, D.J.: Structure-based carbon nanotube sorting by sequence-dependent DNA assembly. Science 302, 1545–1548 (2003)
Tu, X., Manohar, S., Jagota, A., Zheng, M.: DNA sequence motifs for structure-specific recognition and separation of carbon nanotubes. Nature 460, 250–253 (2009)
Harvey, J.D., Jena, P.V., Baker, H.A., Zerze, G.H., Williams, R.M., Galassi, T.V., Roxbury, D., Mittal, J., Heller, D.A.: A carbon nanotube reporter of microRNA hybridization events in vivo. Nat. Biomed. Eng. 1, 0041 (2017)
Kim, K.I., Yoon, S., Chang, J., Lee, S., Cho, H.H., Jeong, S.H., Jo, K., Lee, J.H.: Multifunctional heterogeneous carbon nanotube nanocomposites assembled by DNA-binding peptide anchors. Small 16, e1905821 (2020)
Du, Y., Dong, S.: Nucleic acid biosensors: recent advances and perspectives. Anal. Chem. 89, 189–215 (2017)
Suresh, R.R., Kulandaisamy, A.J., Nesakumar, N., Nagarajan, S., Lee, J.H., Rayappan, J.B.B.: Graphene quantum dots – hydrothermal green synthesis, material characterization and prospects for cervical cancer diagnosis applications: a review. ChemistrySelect 7, e202200655 (2022)
Lim, H., Chang, J., Kim, K.I., Moon, Y., Lee, S., Lee, B., Lee, J.H., Lee, J.: On-chip selection of adenosine aptamer using graphene oxide-coated magnetic nanoparticles. Biomicrofluidics 16, 044102 (2022)
Na, H., Kang, B.-H., Ku, J., Kim, Y., Jeong, K.-H.: On-chip paper electrophoresis for ultrafast screening of infectious diseases. BioChip J. 15, 305–311 (2021)
Tran, T.T.T., Delgado, A., Jeong, S.: Organ-on-a-chip: the future of therapeutic aptamer research? BioChip J. 15, 109–122 (2021)
Yoo, H., Jo, H., Oh, S.S.: Detection and beyond: challenges and advances in aptamer-based biosensors. Mater. Adv. 1, 2663–2687 (2020)
Heo, J.H., Cho, H.H., Lee, J.H.: Surfactant-free nanoparticle–DNA complexes with ultrahigh stability against salt for environmental and biological sensing. Analyst 139, 5936–5944 (2014)
Lee, J.H., Wang, Z., Liu, J., Lu, Y.: Highly sensitive and selective colorimetric sensors for uranyl (UO22+): development and comparison of labeled and label-free DNAzyme-gold nanoparticle systems. J. Am. Chem. Soc. 130, 14217–14226 (2008)
Wang, H., Peng, R., Wang, J., Qin, Z., Xue, L.: Circulating microRNAs as potential cancer biomarkers: the advantage and disadvantage. Clin. Epigenetics 10, 59 (2018)
Kim, D.H., Paek, S.H., Choi, D.Y., Lee, M.K., Park, J.-N., Cho, H.-M., Paek, S.-H.: Real-time monitoring of biomarkers in serum for early diagnosis of target disease. BioChip J. 14, 2–17 (2020)
Paek, S.-H.: Real-time monitoring of biomarkers: current status and future perspectives. BioChip J. 14, 1 (2020)
Mitchell, P.S., Parkin, R.K., Kroh, E.M., Fritz, B.R., Wyman, S.K., Pogosova-Agadjanyan, E.L., Peterson, A., Noteboom, J., O’Briant, K.C., Allen, A., Lin, D.W., Urban, N., Drescher, C.W., Knudsen, B.S., Stirewalt, D.L., Gentleman, R., Vessella, R.L., Nelson, P.S., Martin, D.B., Tewari, M.: Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 105, 10513–10518 (2008)
Weber, J.A., Baxter, D.H., Zhang, S., Huang, D.Y., Huang, K.H., Lee, M.J., Galas, D.J., Wang, K.: The microRNA spectrum in 12 body fluids. Clin Chem. 56, 1733–1741 (2010)
Deng, Z.M., Chen, G.H., Dai, F.F., Liu, S.Y., Yang, D.Y., Bao, A.Y., Cheng, Y.X.: The clinical value of miRNA-21 in cervical cancer: a comprehensive investigation based on microarray datasets. PLoS ONE 17, e0267108 (2022)
Cai, L., Wang, W., Li, X., Dong, T., Zhang, Q., Zhu, B., Zhao, H., Wu, S.: MicroRNA-21-5p induces the metastatic phenotype of human cervical carcinoma cells in vitro by targeting the Von Hippel-Lindau tumor suppressor. Oncol. Lett 15, 5213–5219 (2018)
Kannappan, S., Lee, J.H., Lakshmanakumar, M., Rayappan, J.B.B., Nesakumar, N.: Potential biomarkers for early diagnosis of cervical cancer. In: Rayappan, J.B.B., Lee, J.H. (eds.) Biomarkers and biosensors for cervical cancer diagnosis, pp. 23–46. Springer, Singapore (2021)
Kim, H., Huh, H.J., Park, E., Chung, D.-R., Kang, M.: Multiplex molecular point-of-care test for syndromic infectious diseases. BioChip J. 15, 14–22 (2021)
Torre, L.A., Siegel, R.L., Ward, E.M., Jemal, A.: Global cancer incidence and mortality rates and trends–an update. Cancer Epidemiol. Biomarkers Prev. 25, 16–27 (2016)
Guerrero, J.M., Aguirre, F.S., Mota, M.L., Carrillo, A.: Advances for the development of in vitro immunosensors for multiple sclerosis diagnosis. BioChip J. 15, 205–215 (2021)
Park, S., Eom, K., Kim, J., Bang, H., Wang, H.Y., Ahn, S., Kim, G., Jang, H., Kim, S., Lee, D., Park, K.H., Lee, H.: MiR-9, miR-21, and miR-155 as potential biomarkers for HPV positive and negative cervical cancer. BMC Cancer 17, 658 (2017)
Li, S., Olson, W.K., Lu, X.-J.: Web 3DNA 2.0 for the analysis, visualization, and modeling of 3D nucleic acid structures. Nucleic Acids Res. 47, W26–W34 (2019)
Pettersen, E.F., Goddard, T.D., Huang, C.C., Meng, E.C., Couch, G.S., Croll, T.I., Morris, J.H., Ferrin, T.E.: UCSF ChimeraX: structure visualization for researchers, educators, and developers. Protein Sci. 30, 70–82 (2021)
Choi, Y.K., Kern, N.R., Kim, S., Kanhaiya, K., Afshar, Y., Jeon, S.H., Jo, S., Brooks, B.R., Lee, J., Tadmor, E.B., Heinz, H., Im, W.: CHARMM-GUI nanomaterial modeler for modeling and simulation of nanomaterial systems. J Chem Theory Comput. 18, 479–493 (2022)
Jo, S., Kim, T., Iyer, V.G., Im, W.: CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 29, 1859–1865 (2008)
Frey, J. T. & Doren, D. J. TubeGen 3.4, <http://turin.nss.udel.edu/research/tubegenonline.html> (2011)
Ghosh, S., Patel, N., Chakrabarti, R.: Probing the salt concentration dependent nucleobase distribution in a single-stranded DNA–single-walled carbon nanotube hybrid with molecular dynamics. J. Phys. Chem. B 120, 455–466 (2016)
Van Der Spoel, D., Lindahl, E., Hess, B., Groenhof, G., Mark, A.E., Berendsen, H.J.: GROMACS: fast, flexible, and free. J Comput Chem. 26, 1701–1718 (2005)
Bjelkmar, P., Larsson, P., Cuendet, M.A., Hess, B., Lindahl, E.: Implementation of the CHARMM force field in GROMACS: analysis of protein stability effects from correction maps, virtual interaction sites, and water models. J. Chem. Theory Comput. 6, 459–466 (2010)
Beglov, D., Roux, B.: Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J. Chem. Phys. 100, 9050–9063 (1994)
Jorgensen, W.L., Chandrasekhar, J., Madura, J.D., Impey, R.W., Klein, M.L.: Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983)
Schrodinger, LLC.: The PyMOL Molecular Graphics System, Version 1.8 (2015)
Valdés-Tresanco, M.S., Valdés-Tresanco, M.E., Valiente, P.A., Moreno, E.: gmx_MMPBSA: a new tool to perform end-state free energy calculations with GROMACS. J. Chem. Theory Comput. 17, 6281–6291 (2021)
Jung, S., Cha, M., Park, J., Jeong, N., Kim, G., Park, C., Ihm, J., Lee, J.: dissociation of single-strand DNA: single-walled carbon nanotube hybrids by Watson-Crick base-pairing. J. Am. Chem. Soc. 132, 10964–10966 (2010)
Johnson, R.R., Johnson, A.T.C., Klein, M.L.: Probing the structure of DNA-carbon nanotube hybrids with molecular dynamics. Nano Lett. 8, 69–75 (2008)
Jeng, E.S., Barone, P.W., Nelson, J.D., Strano, M.S.: Hybridization kinetics and thermodynamics of DNA adsorbed to individually dispersed single-walled carbon nanotubes. Small 3, 1602–1609 (2007)
Shankar, A., Mittal, J., Jagota, A.: Binding between DNA and carbon nanotubes strongly depends upon sequence and chirality. Langmuir 30, 3176–3183 (2014)
Johnson, R.R., Johnson, A.T.C., Klein, M.L.: The nature of DNA-base–carbon-nanotube interactions. Small 6, 31–34 (2010)
Acknowledgements
We acknowledge the support received from the National Research Foundation (NRF) of Korea for the Basic Science Research Program (NRF-2019R1A6A1A03033215) and the Korea Basic Science Institute (National Research Facilities and Equipment Center) grant (2020R1A6C103B101) funded by the Ministry of Education. This research was also supported by the National Research Foundation (NRF) of Korea for Bio & Medical Technology Development Program (NRF-2022M3A9G8017220) funded by the Ministry of Science & ICT. The authors are grateful to the Ministry of Human Resource Development, Government of India, for financial support through the Scheme for Promotion of Academic and Research Collaboration (SPARC) project “SPARC/2018-19/P402/SL”. Shrute Kannappan appreciates Sungkyunkwan University for the SKKU Research Matters Fellowship.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declares that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Kannappan, S., Chang, J., Sundharbaabu, P.R. et al. DNA-Wrapped CNT Sensor for Small Nucleic Acid Detection: Influence of Short Complementary Sequence. BioChip J 16, 490–500 (2022). https://doi.org/10.1007/s13206-022-00088-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-022-00088-7