Abstract
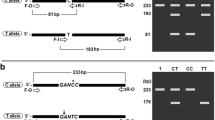
Recently, polymorphisms in the clusterin (CLU) or Apolipoprotein J (ApoJ) gene were reported to be involved in lipid metabolism, atherogenesis, and being associated with the risk of developing Alzheimer’s disease (AD). The influences from genetic variation has not been examined among Koreans. To screen genes with abnormal exon expression profiles, we developed PCR primer pairs for CLU gene. The new primer set can target specific regions of previously sequenced CLU gene. Primers were designed to target eight exon amplicons with flanking regions to optimize PCR amplification. One-hundred samples from clinically diagnosed Korean AD patients were selected for testing. We identified four single nucleotide polymorphisms (SNPs) in CLU through direct sequencing of the PCR products. These included three SNPs (rs7982, rs2279590 and rs3216167) that were previously reported variants in the SNP database (dbSNP), which could be found in NCBI. Interestingly, one new (NEW1) or extremely low-frequency mutation was found in more than one individual among the late-onset AD subjects. Our study suggests that CLU variants may also be an AD susceptibility factor among Koreans. Moreover, the findings of the study can be able to develop for simultaneous identify all of the genotypes of CLU mutation sites by DNA microarray PCR-based in the future.
Similar content being viewed by others
References
Agnishwar, G., Jung, C., Mun, H.Y. & Park, H.G. PCRfree mutation detection of BRCA1 on a zip-code microarray using ligase chain reaction. Journal of Biochemical and Biophysical Method. 70, 897–902 (2008).
Aronow, B.J., Lund, S.D., Brown, T.L., Harmony, J.A. & Witte, D.P. Apolipoprotein J expression at fluid-tissue interfaces: potential role in barrier cytoprotection. Proc. Natl. Acad. Sci. USA. 90, 725–729 (1993).
Braskie, M.N. et al. Common Alzheimer’s disease risk variant within the CLU gene affects white matter microstructure in young adults. Journal of Neuroscienc. 31, 6764–6770 (2011).
Calero, M. et al. Apolipoprotein J (clusterin) and Alzheimer’s disease. Microsc. Res. Tech. 50, 305–315 (2000).
Carrasquillo, M.M., Belbin, O., Hunter, T.A., Ma, L. & Bisceglio, G.D. Replication of CLU, CR1, and PICALM associations with Alzheimer disease. Archives of Neurolog. 67, 961–964 (2010).
Chen, L.H. et al. Polymorphisms of CR1, CLU and PICALM confer susceptibility of Alzheimer’s disease in a southern Chinese population. Neurobiol. Agin. 33, 211–217 (2012).
Daimon, M. et al. Association of the clusterin gene polymorphisms with type 2 diabetes mellitus. Metabolis. 60, 815–822 (2011).
Elias-Sonnenschein, L.S. et al. Genetic Loci Associated with Alzheimer’s Disease and Cerebrospinal Fluid Biomarkers in a Finnish Case-Control Cohort. PLoS One 8, e59676 (2013).
Eva, B., Youn, Y.C., An, S.S.A. & Kim, S.Y. The genetics of Alzheimer’s disease. Clin. Interv. Agin. 9, 535–551 (2014).
Guerreiro, R.J. et al. Genetic variability in CLU and its association with Alzheimer’s disease. PLoS One 5, e9510 (2010).
Harold, D. et al. Genome-wide association study identifies variants at CLU and PICALM associated with Alzheimer’s disease. Nat. Genet. 41, 1088–1093 (2009).
Jun, G. et al. Meta-analysis confirms CR1, CLU, and PICALM as Alzheimer disease risk loci and reveals interactions with APOE genotypes. Arch Neurol. 67, 1473–1484 (2010).
Kamboh, M.I. et al. Association of CLU and PICALM variants with Alzheimer’s disease. Neurobiol. Agin. 33, 518–521 (2012).
Komatsu, M. et al. Genetic association between clusterin polymorphisms and Alzheimer’s disease in a Japanese population. Psychogeriatric. 11, 14–18 (2011).
Lambert, J.C. et al. Genomewide association study identifies variants at CLU and CR1 associated with Alzheimer’s disease. Nat. Genet. 41, 1094–1099 (2009).
Lancaster, T.M. et al. Neural hyperactivation in carriers of the Alzheimer’s risk variant on the clusterin gene. European Neuropsychopharmacolog. 21, 880–884 (2011).
May, P.C. et al. Dynamics of gene expression for a hippocampal glycoprotein elevated in Alzheimer’s disease and in response to experimental lesions in rat. Neuro. 5, 831–839 (1990).
Miyashita, A. et al. SORL1 is genetically associated with late-onset Alzheimer’s disease in Japanese, Koreans and Caucasians. PLoS One 8, e58618 (2013).
Song, J.Y., Park, H.G., Jung, S.O. & Park, J. Diagnosis of HNF-1alpha mutations on a PNA zip-code microarray by single base extension. Nucleic Acids Res. 33, p. e19 (2005).
Swaroop, A., Chew, E.Y., Rickman, C.B. & Abecasis, G.R. Unraveling a multifactorial late-onset disease: from genetic susceptibility to disease mechanisms for age-related macular degeneration. Annu. Rev. Genomics Hum. Genet. 10, 19–43 (2009).
Sweet, R.A. et al. Effect of Alzheimer’s disease risk genes on trajectories of cognitive function in the Cardiovascular Health Study. Am. J. Psychiatr. 169, 954–962 (2012).
Thambisetty, M. et al. Plasma clusterin concentration is associated with longitudinal brain atrophy in mild cognitive impairment. NeuroImag. 59, 212–217 (2012).
Tyagi, S., Bratu, D.P. & Kramer, F.R. Multicolor molecular beacons for allele discrimination. Nat. Biotechnol. 16, 49–53 (1998).
Untergasser, A. et al. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Res. 35, W71–W74 (2007).
Wang, D.G. et al. Large-scale identification, mapping, and genotyping of single-nucleotide polymorphism in the human genome. Scienc. 280, 1077–1082 (1998).
Wu, Z.C., Yu, J.T., Li, Y. & Tan, L. Clusterin in Alzheimer’s disease. Advances in Clinical Chemistr. 56, 155–173 (2012).
Yim, S.C., Park, H.G., Chang, H.N. & Cho, D.Y. Array-based mutation detection of BRCA1 using direct probe/target hybridization. Anal. Biochem. 337, 332–337 (2005).
Yu, J.T. et al. Implication of CLU gene polymorphisms in Chinese patients with Alzheimer’s disease. Clin. Chim. Act. 411, 1516–1519 (2010).
Zabarovsky, E.R. et al. Restriction site tagged (RST) microarrays: a novel technique to study the species composition of complex microbial systems. Nucleic Acids Res. 31, e95 (2003).
Zhang, S. et al. CLU rs2279590 polymorphism contributes to Alzheimer’s disease susceptibility in Caucasian and Asian populations. J. Neural. Transm. 122, 433–439 (2015).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Van Giau, V., An, S.S.A. Optimization of specific multiplex DNA primers to detect variable CLU genomic lesions in patients with Alzheimer’s disease. BioChip J 9, 278–284 (2015). https://doi.org/10.1007/s13206-015-9306-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-015-9306-8