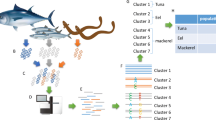
Abstract
Hairtails of the family Trichiuridae are widely distributed in the West Sea, South Sea, and Jeju Island in Korea and form large populations on the continental shelf of the western North Pacific. These fish species are imported from China and several other countries because of the high demand in Korea. However, imported hairtail are difficult to distinguish from domestic hairtail. Thus, we developed a DNA chip that distinguishes three hairtail species from eight countries for quick and simple species identification. Species-specific oligonucleotide probes were designed by sequence analysis of the mitochondrial cytochrome c oxidase subunit I. In this study, we used species-specific probes and a DNA chip system to successfully and rapidly identify three different hairtail species from eight different geographical locations.
Similar content being viewed by others
References
Chakraborty, A., Aranishi, F. & Iwatsuki, Y. Genetic differences among three species of the genus Trichiurus (Perciformes: Trichiuridae) based on mitochondrial DNA analysis. Ichthyol. Res. 53, 93–96 (2006).
Chung, I.H. et al. A DNA microarray for identification of selected Korean birds based on mitochondrial cytochrome c oxidase I gene sequences. Mol. Cells 30, 295–301 (2010).
Hebert, P.D.N., Stoeckle, M.Y., Zemlak, T.S. & Francis, C.M. Identification of birds through DNA barcodes. PLoS Biol. 2, e312 (2004).
Kress, W.J. & Erickson, D.L. DNA barcodes: genes, genomics, and bioinformatics. Proc. Natl. Acad. Sci. 105, 2761–2762 (2008).
Valdez-Moreno, M., Ivanova, N., Elías-Gutiérrez, M., Contreras-Balderas, S. & Hebert, P. Probing diversity in freshwater fishes from Mexico and Guatemala with DNA barcodes. J. Fish Biol. 74, 377–402 (2009).
Ward, R.D., Zemlak, T.S., Innes, B.H., Last, P.R. & Hebert, P.D.N. DNA barcoding Australia’s fish species. Phil. Trans. R. Soc. B. 360, 1847–1857 (2005).
Hebert, P.D.N., Cywinska, A. & Ball, S.L. Biological identifications through DNA barcodes. Phil. Trans. R. Soc. B. 270, 313–321 (2003).
Nijman, V. & Aliabadian, M. Performance of distance-based DNA barcoding in the molecular identification of primates. C. R. Biologies 333, 11–16 (2010).
Rasmussen, R.S., Morrissey, M.T. & Hebert, P.D.N. DNA barcoding of commercially important salmon and trout species (Oncorhynchus and Salmo) from North America. J. Agric. Food Chem. 57, 8379–8385 (2009).
Steinke, D., Zemlak, T.S. & Hebert, P.D.N. Barcoding Nemo: DNA-based identifications for the ornamental fish trade. PLoS ONE 4, e6300 (2009).
Tzeng, C.H. & Chiu, T.S. DNA barcode-based identification of commercially caught cutlassfishes (Family: Trichiuridae) with a phylogenetic assessment. Fish. Res. 176–181 (2012).
Lowenstein, J.H., Amato, G. & Kolokotronis, S.O. The real maccoyii: identifying tuna sushi with DNA barcodes-contrasting characteristic attributes and genetic distances. PLoS ONE 4, e7866 (2009).
Yancy, H.F. et al. Potential use of DNA barcodes in regulatory science: applications of the Regulatory Fish Encyclopedia. J. Food Prot. 71, 210–217 (2008).
Ivanova, N.V., Zemlak, T.S., Hanner, R.H. & Herbert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 7, 544–548 (2007).
Lakra, W. et al. DNA barcoding Indian marine fishes. Mol. Ecol. Resour. 11, 60–71 (2011).
Ward, R., Hanner, R. & Hebert, P. The campaign to DNA barcode all fishes, FISH-BOL. J. Fish Biol. 74, 329–356 (2009).
Kochzius, M. et al. DNA microarrays for identifying fishes. Mar. Biotechnol. 10, 207–217 (2008).
Kochzius, M. et al. Identifying fishes through DNA barcodes and microarrays. PLoS ONE 5, e12620 (2010).
Park, J.Y. et al. A DNA microarray for species identification of cetacean animals in Korean water. BioChip J. 4, 197–203 (2010).
Peytavi, R. et al. Correlation between microarray DNA hybridization efficiency and the position of short capture probe on the target nucleic acid. BioTechniques 39, 89 (2005).
Zhang, L., Hurek, T. & Reinhold-Hurek, B. Position of the fluorescent label is a crucial factor determining signal intensity in microarray hybridizations. Nucleic Acids Research 33, e166–e166 (2005).
Hubert, N. et al. Identifying Canadian freshwater fishes through DNA barcodes. PLoS ONE 3, e2490 (2008).
Smith, P. et al. DNA barcoding highlights a cryptic species of grenadier Macrourus in the Southern Ocean. J. Fish Biol. 78, 355–365 (2011).
Vrijenhoek, R. DNA primers for amplification of mitochondrial cytochrome c oxidase subunit I from diverse metazoan invertebrates. Mol. Mar. Biol. Biotechnol. 3, 294–299 (1994).
Tamura, K., Dudley, J., Nei, M. & Kumar, S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24, 1596–1599 (2007).
Kimura, M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J. Mol. Evol. 16, 111–120 (1980).
Saitou, N. & Nei, M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 4, 406–425 (1987).
Felsenstein, J. Confidence limits on phylogenies: an approach using the bootstrap. Evolution 783–791 (1985).
Author information
Authors and Affiliations
Corresponding author
Additional information
These authors contributed equally to this work.
Rights and permissions
About this article
Cite this article
Park, J.Y., Kim, JH., Kim, EM. et al. Development of a DNA chip to identify the place of origin of hairtail species. BioChip J 7, 136–142 (2013). https://doi.org/10.1007/s13206-013-7206-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-013-7206-8