Abstract
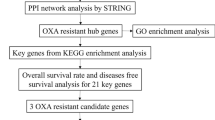
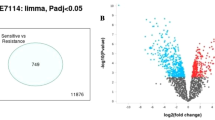
The present study aimed to identify the differentially expressed genes (DEGs) and enriched pathways in docetaxel (DTX) resistant breast cancer cell lines by bioinformatics analysis. The microarray dataset GSE28784 was obtained from gene expression omnibus (GEO) database. The differentially expressed genes (DEGs), gene ontology (GO), and Kyoto Encyclopedia of gene and genome (KEGG) pathway analyses were performed with the help of GEO2R and DAVID tools. Furthermore, the protein–protein interaction (PPI) and hub-gene network of DEGs were constructed using STRING and Cytohubba tools. The prognostic values of hub genes were calculated with the help of the Kaplan–Meier plotter database. From the GEO2R analysis, 222 DEGs were identified of which 120 are upregulated and 102 are downregulated genes. In the PPIs network, five up-regulated genes including CCL2, SPARC, CYR61, F3, and MFGE8 were identified as hub genes. It was observed that low expression of six hub genes CXCL8, CYR61, F3, ICAM1, PLAT, and THBD were significantly correlated with poor overall survival of BC patients in survival analysis. miRNA analysis identified that hsa-mir-16-5p, hsa-mir-335-5p, hsa-mir-124-3p, hsa-mir-20a-5p, and hsa-mir-155-5p are the top 5 interactive miRNAs that are commonly interacting with more hub genes with degree score of greater than five. Additionally, drug-gene interaction analysis was performed to identify drugs which are could potentially elevate/lower the expression levels of hub genes. In summary, the gene-miRNAs-TFs network and subsequent correlation of candidate drugs with hub genes may improve individualized diagnosis and help select appropriate combination therapy for DTX-resistant BC in the future.






Similar content being viewed by others
Data availability
Data could be provided as per request.
References
Brown I, Shalli K, McDonald SL, Moir SE, Hutcheon AW, Heys SD, Schofield AC (2004) Reduced expression of p27 is a novel mechanism of docetaxel resistance in breast cancer cells. Breast Cancer Res 6(5):R601. https://doi.org/10.1186/bcr918
Chan HJ, Li H, Liu Z, Yuan Y-C, Mortimer J, Chen S (2015) SERPINA1 is a direct estrogen receptor target gene and a predictor of survival in breast cancer patients. Oncotarget 6(28):25815. https://doi.org/10.18632/oncotarget.4441
Cocco E, Varughese J, Buza N, Bellone S, Lin K-Y, Bellone M, Todeschini P, Silasi D-A, Azodi M, Schwartz PE (2011) Tissue factor expression in ovarian cancer: implications for immunotherapy with hI-con1, a factor VII-IgGF c chimeric protein targeting tissue factor. Clin Exp Metastasis 28(7):689–700. https://doi.org/10.1007/s10585-011-9401-0
Corte MD, Vérez P, Rodríguez JC, Roibás A, Domínguez ML, Lamelas ML, Vázquez J, Muñiz JLG, Allende MT, González LO (2005) Tissue-type plasminogen activator (tPA) in breast cancer: relationship with clinicopathological parameters and prognostic significance. Breast Cancer Res Treat 90(1):33–40. https://doi.org/10.1007/s10549-004-2624-x
Das AM, Bolkestein M, van der Klok T, Ophuis CMO, Vermeulen CE, Rens JA, Dinjens WN, Atmodimedjo PN, Verhoef C, Koljenović S (2016) Tissue inhibitor of metalloproteinase-3 (TIMP3) expression decreases during melanoma progression and inhibits melanoma cell migration. Eur J Cancer 66:34–46. https://doi.org/10.1016/j.ejca.2016.06.020
de Groote ML, Kazemier HG, Huisman C, van der Gun BT, Faas MM, Rots MG (2014) Upregulation of endogenous ICAM-1 reduces ovarian cancer cell growth in the absence of immune cells. Int J Cancer 134(2):280–290. https://doi.org/10.1002/ijc.28375
Egawa C, Miyoshi Y, Takamura Y, Taguchi T, Tamaki Y, Noguchi S (2001) Decreased expression of BRCA2 mRNA predicts favorable response to docetaxel in breast cancer. Int J Cancer 95(4):255–259. https://doi.org/10.1002/1097-0215(20010720)95:4%3c255::aid-ijc1043%3e3.0.co;2-o
Feng H, Cheung AN, Xue W-C, Wang Y, Wang X, Fu S, Wang Q, Ngan HY, Tsao S-W (2004) Down-regulation and promoter methylation of tissue inhibitor of metalloproteinase 3 in choriocarcinoma. Gynecol Oncol 94(2):375–382. https://doi.org/10.1016/j.ygyno.2004.04.019
Ferlay J, Steliarova-Foucher E, Lortet-Tieulent J, Rosso S, Coebergh J-WW, Comber H, Forman D, Bray F (2013) Cancer incidence and mortality patterns in Europe: estimates for 40 countries in 2012. Eur J Cancer 49(6):1374–1403. https://doi.org/10.1016/j.ejca.2012.12.027
Geng W, Song H, Zhao Q, Dong K, Pu Q, Gao H, Lv Y (2020) miR-520h stimulates drug resistance to paclitaxel by targeting the OTUD3-PTEN axis in breast cancer. Biomed Res Int. https://doi.org/10.1155/2020/9512793
Han X-g, Mo H-m, Liu X-q, Li Y, Du L, Qiao H, Fan Q-m, Zhao J, Zhang S-h, Tang T-t (2018) TIMP3 overexpression improves the sensitivity of osteosarcoma to cisplatin by reducing IL-6 production. Front Genet 9:135. https://doi.org/10.3389/fgene.2018.00135
Hedges JC, Singer CA, Gerthoffer WT (2000) Mitogen-activated protein kinases regulate cytokine gene expression in human airway myocytes. Am J Respir Cell Mol Biol 23(1):86–94. https://doi.org/10.1165/ajrcmb.23.1.4014
Hu Q, Chen W-x, Zhong S-l, Zhang J-y, Ma T-f, Ji H, Lv M-m, Tang J-h, Zhao J-h (2014) MicroRNA-452 contributes to the docetaxel resistance of breast cancer cells. Tumor Biol 35(7):6327–6334. https://doi.org/10.1007/s13277-014-1834-z
Huang P, Li F, Li L, You Y, Luo S, Dong Z, Gao Q, Wu S, Brünner N, Stenvang J (2018) lncRNA profile study reveals the mRNAs and lncRNAs associated with docetaxel resistance in breast cancer cells. Sci Rep 8(1):1–15. https://doi.org/10.1038/s41598-018-36231-4
Kastl L, Brown I, Schofield AC (2012) miRNA-34a is associated with docetaxel resistance in human breast cancer cells. Breast Cancer Res Treat 131(2):445–454. https://doi.org/10.1007/s10549-011-1424-3
Kwon C, Park H, Lee J, Kim H, Jeon T, Jo H, Kim D, Kim G, Park D (2014) Serpin peptidase inhibitor clade A member 1 is a biomarker of poor prognosis in gastric cancer. Br J Cancer 111(10):1993–2002. https://doi.org/10.1038/bjc.2014.490
Leonard GD, Fojo T, Bates SE (2003) The role of ABC transporters in clinical practice. Oncologist 8(5):411–424. https://doi.org/10.1634/theoncologist.8-5-411
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20. https://doi.org/10.1016/j.cell.2004.12.035
Liu Q, Li A, Tian Y, Wu JD, Liu Y, Li T, Chen Y, Han X, Wu K (2016) The CXCL8-CXCR1/2 pathways in cancer. Cytokine Growth Fact Rev 31:61–71. https://doi.org/10.1016/j.cytogfr.2016.08.002
Masson D, Rioux-Leclercq N, Fergelot P, Jouan F, Mottier S, Théoleyre S, Bach-Ngohou K, Patard J-J, Denis MG (2010) Loss of expression of TIMP3 in clear cell renal cell carcinoma. Eur J Cancer 46(8):1430–1437. https://doi.org/10.1016/j.ejca.2010.01.009
Mattingly CJ, Colby GT, Forrest JN, Boyer JL (2003) The comparative toxicogenomics database (CTD). Environ Health Perspect 111(6):793–795
Miyoshi Y, Ando A, Takamura Y, Taguchi T, Tamaki Y, Noguchi S (2002) Prediction of response to docetaxel by CYP3A4 mRNA expression in breast cancer tissues. Int J Cancer 97(1):129–132. https://doi.org/10.1002/ijc.1568
Organization WH (2016) World health statistics 2016: monitoring health for the SDGs sustainable development goals. World Health Organization
Rivoltini L, Cattoretti G, Arienti F, Mastroianni A, Melani C, Colombo MP, Parmiani G (1991) The high lysability by lak cells of colon-carcinoma cells resistant to doxorubicin is associated with a high expression of ICAM-1, LFA-3, NCA and a less-differentiated phenotype. Int J Cancer 47(5):746–754. https://doi.org/10.1002/ijc.2910470521
Schröder C, Witzel I, Müller V, Krenkel S, Wirtz RM, Jänicke F, Schumacher U, Milde-Langosch K (2011) Prognostic value of intercellular adhesion molecule (ICAM)-1 expression in breast cancer. J Cancer Res Clin 137(8):1193–1201. https://doi.org/10.3389/fimmu.2023.1176647
Shi Z, Yang W-M, Chen L-P, Yang D-H, Zhou Q, Zhu J, Chen J-J, Huang R-C, Chen Z-S, Huang R-P (2012) Enhanced chemosensitization in multidrug-resistant human breast cancer cells by inhibition of IL-6 and IL-8 production. Breast Cancer Res Treat 135(3):737–747. https://doi.org/10.1007/s10549-012-2196-0
Song B, Wang C, Liu J, Wang X, Lv L, Wei L, Xie L, Zheng Y, Song X (2010) MicroRNA-21 regulates breast cancer invasion partly by targeting tissue inhibitor of metalloproteinase 3 expression. J Exp Clin Cancer Res 29(1):1–8. https://doi.org/10.1186/1756-9966-29-29
Tsai S-T, Wang P-J, Liou N-J, Lin P-S, Chen C-H, Chang W-C (2015) ICAM1 is a potential cancer stem cell marker of esophageal squamous cell carcinoma. PLoS One 10(11):e0142834. https://doi.org/10.1371/journal.pone.0142834
Ueno T, Toi M, Koike M, Nakamura S, Tominaga T (2000) Tissue factor expression in breast cancer tissues: its correlation with prognosis and plasma concentration. Br J Cancer 83(2):164–170. https://doi.org/10.1054/bjoc.2000.1272
Vrana JA, Stang MT, Grande JP, Getz MJ (1996) Expression of tissue factor in tumor stroma correlates with progression to invasive human breast cancer: paracrine regulation by carcinoma cellderived members of the transforming growth factor β family. Cancer Res 56(21):5063–5070
Wang X, Pei X, Guo G, Qian X, Dou D, Zhang Z, Xu X, Duan X (2020) Exosome-mediated transfer of long noncoding RNA H19 induces doxorubicin resistance in breast cancer. J Cell Physiol. https://doi.org/10.1002/jcp.29585
Wolff B, Burns AR, Middleton J, Rot A (1998) Endothelial cell “memory” of inflammatory stimulation: human venular endothelial cells store interleukin 8 in Weibel-Palade bodies. J Exp Med 188(9):1757–1762. https://doi.org/10.1084/jem.188.9.1757
Wu W, Wang Q, Yin F, Yang Z, Zhang W, Gabra H, Li L (2016) Identification of proteomic and metabolic signatures associated with chemoresistance of human epithelial ovarian cancer. Int J Oncol 49(4):1651–1665. https://doi.org/10.3892/ijo.2016.3652
Yang Y, Yee D (2012) Targeting insulin and insulin-like growth factor signaling in breast cancer. J Mammary Gland Biol Neoplasia 17:251–261. https://doi.org/10.1007/s10911-012-9268-y
Zhang Y, Wang Y, Wei Y, Li M, Yu S, Ye M, Zhang H, Chen S, Liu W, Zhang J (2015) MiR-129-3p promotes docetaxel resistance of breast cancer cells via CP110 inhibition. Sci Rep 5:15424. https://doi.org/10.1038/srep15424
Zhang X, Zhong S, Xu Y, Yu D, Ma T, Chen L, Zhao Y, Chen X, Yang S, Wu Y (2016) MicroRNA-3646 contributes to docetaxel resistance in human breast cancer cells by GSK-3β/β-catenin signaling pathway. PLoS One 11(4):e0153194. https://doi.org/10.1371/journal.pone.0153194
Zhang C, Wang J, Zhang J, Qu H, Tang X (2020) LINC00461 overexpression can induce docetaxel resistance in breast cancer by interacting with miR-411-5p. Onco Targets Ther 13:5551–5562. https://doi.org/10.2147/OTT.S247776
Zheng Y, Yang J, Qian J, Qiu P, Hanabuchi S, Lu Y, Wang Z, Liu Z, Li H, He J (2013) PSGL-1/selectin and ICAM-1/CD18 interactions are involved in macrophage-induced drug resistance in myeloma. Leukemia 27(3):702–710. https://doi.org/10.1038/leu.2012.272
Acknowledgements
Baddipadige Raju and Gera Narendra would like to acknowledge the Indian Council of Medical Research (ICMR), New Delhi for providing a Senior Research Fellowship (SRF).
Funding
This work was supported by the Indian Council of Medical Research (ICMR), New Delhi; Sanction No. ISRM/12(10)/2019.
Author information
Authors and Affiliations
Contributions
Baddipadige Raju: Concept, Designing of Work, Original Manuscript Writing, Editing, Software usage, Analysis, and Interpretation of Data. Gera Narendra: Designing of work, Software usage, and Data interpretation. Himanshu Verma: Data interpretation, Manuscript Editing. Om Silakari: Designing of work, Editing, and Proofreading.
Corresponding author
Ethics declarations
Conflict of interest
Authors declared no conflict of interest.
Ethical statement
This article does not contain any studies with human participants performed by any of the authors.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Raju, B., Narendra, G., Verma, H. et al. Identification of chemoresistance associated key genes-miRNAs-TFs in docetaxel resistant breast cancer by bioinformatics analysis. 3 Biotech 14, 128 (2024). https://doi.org/10.1007/s13205-024-03971-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-024-03971-2