Abstract
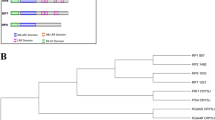
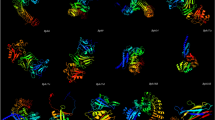
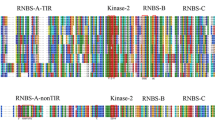
In the current study, gene network analysis revealed five novel disease-resistance proteins against bacterial leaf blight (BB) and rice blast (RB) diseases caused by Xanthomonas oryzae pv. oryzae (Xoo) and Magnaporthe oryzae (M. oryzae), respectively. In silico modeling, refinement, and model quality assessment were performed to predict the best structures of these five proteins and submitted to ModelArchive for future use. An in-silico annotation indicated that the five proteins functioned in signal transduction pathways as kinases, phospholipases, transcription factors, and DNA-modifying enzymes. The proteins were localized in the nucleus and plasma membrane. Phylogenetic analysis showed the evolutionary relation of the five proteins with disease-resistance proteins (XA21, OsTRX1, PLD, and HKD-motif-containing proteins). This indicates similar disease-resistant properties between five unknown proteins and their evolutionary-related proteins. Furthermore, gene expression profiling of these proteins using public microarray data showed their differential expression under Xoo and M. oryzae infection. This study provides an insight into developing disease-resistant rice varieties by predicting novel candidate resistance proteins, which will assist rice breeders in improving crop yield to address future food security through molecular breeding and biotechnology.




Similar content being viewed by others
Data availability
The datasets generated during the current study are available in the ModelArchive repository with IDs ma-u68j8, ma-yfta8, ma-xdomw, ma-n3mnn, and ma-y1dh3.
References
Almagro Armenteros JJ, Sønderby CK, Sønderby SK, Nielsen H, Winther O (2017) DeepLoc: prediction of protein subcellular localization using deep learning. Bioinformatics (Oxf Engl) 33(21):3387–3395. https://doi.org/10.1093/bioinformatics/btx431
Bakade R, Ingole KD, Deshpande S, Pal G, Patil SS, Bhattacharjee S, Prasannakumar MK, Ramu VS (2021) Comparative transcriptome analysis of rice resistant and susceptible genotypes to Xanthomonas oryzae pv. oryzae identifies novel genes to control bacterial leaf blight. Mol Biotechnol 63(8):719–731. https://doi.org/10.1007/S12033-021-00338-3/FIGURES/6
Bargmann BOR, Munnik T (2006) The role of phospholipase D in plant stress responses. Curr Opin Plant Biol 9(5):515–522. https://doi.org/10.1016/J.PBI.2006.07.011
Blum M, Chang HY, Chuguransky S, Grego T, Kandasaamy S, Mitchell A, Nuka G, Paysan-Lafosse T, Qureshi M, Raj S, Richardson L, Salazar GA, Williams L, Bork P, Bridge A, Gough J, Haft DH, Letunic I, Marchler-Bauer A et al (2021) The InterPro protein families and domains database: 20 years on. Nucleic Acids Res 49(D1):D344–D354. https://doi.org/10.1093/NAR/GKAA977
Bteman A, Martin MJ, Orchard S, Magrane M, Agivetova R, Ahmad S, Alpi E, Bowler-Barnett EH, Britto R, Bursteinas B, Bye-A-Jee H, Coetzee R, Cukura A, da Silva A, Denny P, Dogan T, Ebenezer TG, Fan J, Castro LG et al (2021) UniProt: the universal protein knowledgebase in 2021. Nucleic Acids Res 49(D1):D480–D489. https://doi.org/10.1093/NAR/GKAA1100
Calle García J, Guadagno A, Paytuvi-Gallart A, Saera-Vila A, Amoroso CG, D’esposito D, Andolfo G, AieseCigliano R, Sanseverino W, Ercolano MR, Sanseverino W (2022) PRGdb 4.0: an updated database dedicated to genes involved in plant disease resistance process. Nucleic Acids Res 50(D1):D1483–D1490. https://doi.org/10.1093/NAR/GKAB1087
Chithrashree, Udayashankar AC, Chandra Nayaka S, Reddy MS, Srinivas C (2011) Plant growth-promoting rhizobacteria mediate induced systemic resistance in rice against bacterial leaf blight caused by Xanthomonas oryzae pv. oryzae. Biol Control 59(2):114–122. https://doi.org/10.1016/J.BIOCONTROL.2011.06.010
Choi SC, Lee S, Kim SR, Lee YS, Liu C, Cao X, An G (2014) Trithorax group protein Oryza sativa trithorax1 controls flowering time in rice via interaction with early heading date3. Plant Physiol 164(3):1326–1337. https://doi.org/10.1104/PP.113.228049
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci Publ Protein Soc 2(9):1511–1519. https://doi.org/10.1002/PRO.5560020916
de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N (2006) ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34(suppl_2):W362–W365. https://doi.org/10.1093/NAR/GKL124
Diaz-Granados A, Petrescu AJ, Goverse A, Smant G (2016) SPRYSEC effectors: a versatile protein-binding platform to disrupt plant innate immunity. Front Plant Sci. https://doi.org/10.3389/fpls.2016.01575
Eisenberg D, Lüthy R, Bowie JU (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404. https://doi.org/10.1016/S0076-6879(97)77022-8
Fariselli P, Riccobelli P, Casadio R (1999) Role of evolutionary information in predicting the disulfide-bonding state of cysteine in proteins. Proteins. https://doi.org/10.1002/(SICI)1097-0134(19990815)36:3
Gasteiger E, Gattiker A, Hoogland C, Ivanyi I, Appel RD, Bairoch A (2003) ExPASy: the proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res 31(13):3784–3788. https://doi.org/10.1093/NAR/GKG563
Gowda M, Shirke MD, Mahesh HB, Chandarana P, Rajamani A, Chattoo BB (2015) Genome analysis of rice-blast fungus Magnaporthe oryzae field isolates from southern India. Genomics Data 5:284–291. https://doi.org/10.1016/J.GDATA.2015.06.018
He Z, Xin Y, Wang C, Yang H, Xu Z, Cheng J, Li Z, Ye C, Yin H, Xie Z, Jiang N, Huang J, Xiao J, Tian B, Liang Y, Zhao K, Peng J (2022) Genomics-assisted improvement of super high-yield hybrid rice variety “Super 1000” for resistance to bacterial blight and blast diseases. Front Plant Sci 13:1430. https://doi.org/10.3389/FPLS.2022.881244/BIBTEX
Kanehisa M, Subramaniam (2002) The KEGG database. Novartis Found Symp 247:91–103. https://doi.org/10.1002/0470857897.CH8
Klasberg S, Bitard-Feildel T, Mallet L (2016) Computational identification of novel genes: current and future perspectives. Bioinform Biol Insights 10:121–131. https://doi.org/10.4137/BBI.S39950
Kumar S, Meshram S, Sinha A (2017).Bacterial diseases in rice and their eco-friendly management. www.tjprc.org
Kumar S, Zaharin N, Nadarajah K (2018) In silico identification of resistance and defense related genes for bacterial leaf blight (BLB) in rice. J Pure Appl Microbiol 12(4):1867–1876. https://doi.org/10.22207/JPAM.12.4.22
Kumar A, Kumar R, Shukla P, Singh S, Alam M, Singh DK (2022) Update on cloning and molecular characterization of bacterial blight resistance genes in rice. In: Shukla P, Kumar A, Kumar R, Pandey MK (eds) Molecular response and genetic engineering for stress in plants, vol 2. Biotic stress. IOP Publishing, UK, pp 71–715
Laskowski RA, MacArthur MW, Moss DS, Thornton JM (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 26(2):283–291. https://doi.org/10.1107/S0021889892009944
Lee GR, Won J, Heo L, Seok C (2019) GalaxyRefine2: simultaneous refinement of inaccurate local regions and overall protein structure. Nucleic Acids Res 47(W1):W451–W455. https://doi.org/10.1093/NAR/GKZ288
Li W, Zhu Z, Chern M, Yin J, Yang C, Ran L, Cheng M, He M, Wang K, Wang J, Zhou X, Zhu X, Chen Z, Wang J, Zhao W, Ma B, Qin P, Chen W, Wang Y et al (2017) A natural allele of a transcription factor in rice confers broad-spectrum blast resistance. Cell 170(1):114-126.e15. https://doi.org/10.1016/J.CELL.2017.06.008
Lin F, Lazarus EZ, Rhee SY (2020) QTG-Finder2: a generalized machine-learning algorithm for prioritizing QTL causal genes in plants. G3 Genes Genomes Genet 10(7):2411–2421. https://doi.org/10.1534/g3.120.401122
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS, Thanki N, Yamashita RA, Yang M, Zhang D, Zheng C, Lanczycki CJ, Marchler-Bauer A (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48(D1):D265–D268. https://doi.org/10.1093/NAR/GKZ991
Martin EC, Sukarta OCA, Spiridon L, Grigore LG, Constantinescu V, Tacutu R, Goverse A, Petrescu AJ (2020) LRRpredictor—a new LRR motif detection method for irregular motifs of plant NLR proteins using an ensemble of classifiers. Genes 11(3):286. https://doi.org/10.3390/GENES11030286
McGuffin LJ, Bryson K, Jones DT (2000) The PSIPRED protein structure prediction server. Bioinformatics 16(4):404–405. https://doi.org/10.1093/BIOINFORMATICS/16.4.404
Mi J, Yang D, Chen Y, Jiang J, Mou H, Huang J, Ouyang Y, Mou T (2018) Accelerated molecular breeding of a novel P/TGMS line with broad-spectrum resistance to rice blast and bacterial blight in two-line hybrid rice. Rice 11(1):1–12. https://doi.org/10.1186/S12284-018-0203-8/TABLES/
Moreno P, Fexova S, George N, Manning JR, Miao Z, Mohammed S, Muñoz-Pomer A, Fullgrabe A, Bi Y, Bush N, Iqbal H, Kumbham U, Solovyev A, Zhao L, Prakash A, García-Seisdedos D, Kundu DJ, Wang S, Walzer M et al (2022) Expression atlas update: gene and protein expression in multiple species. Nucleic Acids Res 50(D1):D129–D140. https://doi.org/10.1093/nar/gkab1030
Pagni M, Ioannidis V, Cerutti L, Zahn-Zabal M, Jongeneel CV, Hau J, Martin O, Kuznetsov D, Falquet L (2007) MyHits: improvements to an interactive resource for analyzing protein sequences. Nucleic Acids Res 35(suppl_2):W433–W437. https://doi.org/10.1093/NAR/GKM352
Park CJ, Ronald PC (2012) Cleavage and nuclear localization of the rice XA21 immune receptor. Nat Commun 3(1):1–6. https://doi.org/10.1038/ncomms1932
Pradhan M, Bastia D, Samal KC, Dash M, Sahoo JP (2023) Pyramiding resistance genes for bacterial leaf blight (Xanthomonas oryzae pv. Oryzae) into the popular rice variety, Pratikshya through marker assisted backcrossing. Mol Biol Rep. https://doi.org/10.1007/S11033-023-08805-7/TABLES/6
Qi Z, Du Y, Yu J, Zhang R, Yu M, Cao H, Song T, Pan X, Liang D, Liu Y (2023) molecular detection and analysis of blast resistance genes in rice main varieties in Jiangsu Province, China. Agronomy 13(1):157. https://doi.org/10.3390/AGRONOMY13010157/S1
Rea S, Eisenhaber F, O’Carroll D, Strahl BD, Sun ZW, Schmid M, Opravil S, Mechtier K, Ponting CP, Allis CD, Jenuwein T (2000) Regulation of chromatin structure by site-specific histone H3 methyltransferases. Nature 406(6796):593–599. https://doi.org/10.1038/35020506
Schrödinger L, DeLano W (2020) PyMOL. Source: http://www.pymol.org/pymol.. Accessed 2 Jun 2022
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/GR.1239303
Sharma D, Verma N, Pandey C, Verma D, Bhagat PK, Noryang S, Singh K, Tayyeba S, Banerjee G, Sinha AK (2020) MAP kinase as regulators for stress responses in plants. In: Protein kinases and stress signaling in plants. https://doi.org/10.1002/9781119541578.ch15
Sheik SS, Sundararajan P, Hussain ASZ, Sekar K (2002) Ramachandran plot on the web. Bioinformatics 18(11):1548–1549. https://doi.org/10.1093/BIOINFORMATICS/18.11.1548
Shumayla S, Sharma S, Kumar R, Mendu V, Singh K, Upadhyay SK (2016) Genomic dissection and expression profiling revealed functional divergence in Triticum aestivum leucine rich repeat receptor like kinases (TaLRRKs). Front Plant Sci. https://doi.org/10.3389/fpls.2016.01374
Singh BK, Trivedi P (2017) Microbiome and the future for food and nutrient security. Microb Biotechnol 10(1):50. https://doi.org/10.1111/1751-7915.12592
Singh D, Dhiman VK, Pandey H, Dhiman VK, Pandey D (2022) Crosstalk between salicylic acid and auxins, cytokinins, and gibberellins under biotic. Stress. https://doi.org/10.1007/978-3-031-05427-3_11
Snel B, Lehmann G, Bork P, Huynen MA (2000) STRING: a web-server to retrieve and display the repeatedly occurring neighbourhood of a gene. Nucleic Acids Res 28(18):3442–3444. https://doi.org/10.1093/NAR/28.18.3442
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Von Mering C (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery ingenome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/NAR/GKY1131
Törönen P, Holm L (2022) PANNZER-A practical tool for protein function prediction. Protein Sci Publ Protein Soc 31(1):118–128. https://doi.org/10.1002/PRO.4193
Tusnády GE, Simon I (2001) The HMMTOP transmembrane topology prediction server. Bioinformatics 17(9):849–850. https://doi.org/10.1093/BIOINFORMATICS/17.9.849
Wang Y, Zhao JM, Zhang LX, Wang P, Wang SW, Wang H, Wang XX, Liu Z, Zheng WJ (2016) Analysis of the diversity and function of the alleles of the rice blast resistance genes Piz-t, Pita and Pik in 24 rice cultivars. J Integr Agric 15(7):1423–1431. https://doi.org/10.1016/S2095-3119(15)61207-2
Wang L, Zhao L, Zhang X, Zhang Q, Jia Y, Wang G, Li S, Tian D, Li WH, Yang S (2019) Large-scale identification and functional analysis of NLR genes in blast resistance in the Tetep rice genome sequence. Proc Natl Acad Sci U S A 116(37):18479–18487. https://doi.org/10.1073/pnas.1910229116
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, De Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46(W1):W296–W303. https://doi.org/10.1093/NAR/GKY427
Wilkinson L (2011) ggplot2: elegant graphics for data analysis by WICKHAM, H. Biometrics, 67(2). https://doi.org/10.1111/j.1541-0420.2011.01616.x
Wilkinson SP, Davy SK (2018) phylogram: an R package for phylogenetic analysis with nested lists. J Open Source Softw 3:790. https://doi.org/10.21105/joss.00790
Xu D, Zhang Y (2011) Improving the physical realism and structural accuracy of protein models by a two-step atomic-level energy minimization. Biophys J 101(10):2525–2534. https://doi.org/10.1016/J.BPJ.2011.10.024
Zhai K, Deng Y, Liang D, Tang J, Liu J, Yan B, Yin X, Lin H, Chen F, Yang D, Xie Z, Liu JY, Li Q, Zhang L, He Z (2019) RRM transcription factors interact with NLRs and regulate broad-spectrum blast resistance in rice. Mol Cell 74(5):996-1009.e7. https://doi.org/10.1016/J.MOLCEL.2019.03.013
Zhang ZL, Xie Z, Zou X, Casaretto J, Ho THD, Shen QJ (2004) A rice WRKY gene encodes a transcriptional repressor of the gibberellin signaling pathway in aleurone cells 1. Plant Physiol 134(4):1500–1513. https://doi.org/10.1104/pp.103.034967
Zhang J, Chen L, Fu C, Wang L, Liu H, Cheng Y, Li S, Deng Q, Wang S, Zhu J, Liang Y, Li P, Zheng A (2017) Comparative transcriptome analyses of gene expression changes triggered by Rhizoctonia solani AG1 IA infection in resistant and susceptible rice varieties. Front Plant Sci 8:1422. https://doi.org/10.3389/FPLS.2017.01422/BIBTEX
Zhou L, Feng T, Xu S, Gao F, Lam TT, Wang Q, Wu T, Huang H, Zhan L, Li L, Guan Y, Dai Z, Yu G (2022) ggmsa: a visual exploration tool for multiple sequence alignment and associated data. Brief Bioinf 23(4):1–12. https://doi.org/10.1093/BIB/BBAC222
Acknowledgements
PY acknowledges the support from seed grant (project number I/SEED/PY/20200037) funded by the Indian Institute of Technology, Jodhpur, India; VD is thankful for the financial support from the Ministry of Education (MoE), India; and RSS (file number: 09/1125(0019)/2021-EMR-I) is financially supported by the CSIR-NET fellowship.
Funding
VD collected study data, performed primary analysis, and designed the working strategy; SB carried out basic data analysis and contributed to manuscript writing; RSS performed functional enrichment; PY conceptualized and supervised the study; AS edited the manuscript and provided inputs; all authors contributed to writing the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Authors declare that there are no competing financial interests.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Dhiman, V., Biswas, S., Shekhawat, R.S. et al. In silico characterization of five novel disease-resistance proteins in Oryza sativa sp. japonica against bacterial leaf blight and rice blast diseases. 3 Biotech 14, 48 (2024). https://doi.org/10.1007/s13205-023-03893-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03893-5