Abstract
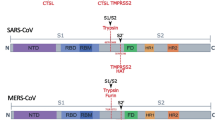
The spike (S) glycoprotein of the SARS-CoV-2 virus binds to the host cell receptor and promotes the virus’s entry into the target host cell. This interaction is primed by host cell proteases like furin and TMPRSS2, which act at the S1/S2 and S2´ cleavage sites, respectively. Both cleavage sites have serine or proline residues flanking either the single or polybasic region and were found to be conserved in coronaviruses. Unravelling the effects of these conserved residues on the virus entry and infectivity might facilitate the development of novel therapeutics. Here, we have investigated the role of the conserved serine and proline residues in the SARS-CoV-2 spike mediated entry, fusogenicity, and viral infectivity by using the HIV-1/spike-based pseudovirus system. A conserved serine residue mutation to alanine (S2´S-A) at the S2´ cleavage site resulted in the complete loss of spike cleavage. Exogenous treatment with trypsin or overexpression of TMPRSS2 protease could not rescue the loss of spike cleavage and biological activity. The S2´S-A mutant showed no significant responses against E-64d, TMPRSS2 or other relevant inhibitors. Taken together, serine at the S2´ site in the spike protein was indispensable for spike protein cleavage and virus infectivity. Thus, novel interventions targeting the conserved serine at the S2´ cleavage site should be explored to reduce severe disease caused by SARS-CoV-2-and novel emerging variants.






Similar content being viewed by others
Data availability
The data supporting the findings of this study are available from the corresponding author upon request.
References
Adedeji AO, Severson W, Jonsson C et al (2013) Novel inhibitors of severe acute respiratory syndrome coronavirus entry that act by three distinct mechanisms. J Virol 87:8017–8028. https://doi.org/10.1128/JVI.00998-13
Alaofi AL, Shahid M (2021) Mutations of SARS-CoV-2 RBD may alter its molecular structure to improve its infection efficiency. Biomolecules. https://doi.org/10.3390/biom11091273
Amitai A (2021) Viral surface geometry shapes influenza and coronavirus spike evolution through antibody pressure. PLoS Comput Biol 17:e1009664. https://doi.org/10.1371/journal.pcbi.1009664
Arora P, Sidarovich A, Krüger N et al (2021) B.1.617.2 enters and fuses lung cells with increased efficiency and evades antibodies induced by infection and vaccination. Cell Rep 37:109825. https://doi.org/10.1016/j.celrep.2021.109825
Belouzard S, Chu VC, Whittaker GR (2009) Activation of the SARS coronavirus spike protein via sequential proteolytic cleavage at two distinct sites. Proc Natl Acad Sci U S A 106:5871–5876. https://doi.org/10.1073/pnas.0809524106
Belouzard S, Millet JK, Licitra BN, Whittaker GR (2012) Mechanisms of coronavirus cell entry mediated by the viral spike protein. Viruses 4:1011–1033. https://doi.org/10.3390/v4061011
Bestle D, Heindl MR, Limburg H et al (2020) TMPRSS2 and furin are both essential for proteolytic activation of SARS-CoV-2 in human airway cells. Life Sci Alliance. https://doi.org/10.26508/lsa.202000786
Bosch BJ, van der Zee R, de Haan CAM, Rottier PJM (2003) The coronavirus spike protein is a class I virus fusion protein: structural and functional characterization of the fusion core complex. J Virol 77:8801–8811. https://doi.org/10.1128/jvi.77.16.8801-8811.2003
Chen Z, Du R, Galvan Achi JM et al (2021) SARS-CoV-2 cell entry and targeted antiviral development. Acta Pharm Sin B 11:3879–3888. https://doi.org/10.1016/j.apsb.2021.05.007
Chu H, Hu B, Huang X et al (2021) Host and viral determinants for efficient SARS-CoV-2 infection of the human lung. Nat Commun 12:134. https://doi.org/10.1038/s41467-020-20457-w
Coronaviridae Study Group of the International Committee on Taxonomy of Viruses (2020) The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2. Nat Microbiol 5:536–544. https://doi.org/10.1038/s41564-020-0695-z
Essalmani R, Jain J, Susan-Resiga D et al (2022) Distinctive roles of furin and TMPRSS2 in SARS-CoV-2 infectivity. J Virol 96:e0012822. https://doi.org/10.1128/jvi.00128-22
Ghimire D, Han Y, Lu M (2022) Structural plasticity and immune evasion of SARS-CoV-2 spike variants. Viruses. https://doi.org/10.3390/v14061255
Hatmal MM, Alshaer W, Al-Hatamleh MAI et al (2020) Comprehensive structural and molecular comparison of spike proteins of SARS-CoV-2, SARS-CoV and MERS-CoV, and their interactions with ACE2. Cells. https://doi.org/10.3390/cells9122638
Hoffmann M, Kleine-Weber H, Pöhlmann S (2020a) A multibasic cleavage site in the spike protein of SARS-CoV-2 is essential for infection of human lung cells. Mol Cell 78:779-784.e5. https://doi.org/10.1016/j.molcel.2020.04.022
Hoffmann M, Kleine-Weber H, Schroeder S et al (2020b) SARS-CoV-2 Cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 181:271-280.e8. https://doi.org/10.1016/j.cell.2020.02.052
Hörnich BF, Großkopf AK, Schlagowski S et al (2021) SARS-CoV-2 and SARS-CoV spike-mediated cell-cell fusion differ in their requirements for receptor expression and proteolytic activation. J Virol. https://doi.org/10.1128/JVI.00002-21
Huang Y, Yang C, Xu X-F et al (2020) Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacol Sin 41:1141–1149. https://doi.org/10.1038/s41401-020-0485-4
Jackson CB, Farzan M, Chen B, Choe H (2022) Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol 23:3–20. https://doi.org/10.1038/s41580-021-00418-x
Khatri R, Siddqui G, Sadhu S et al (2023) Intrinsic D614G and P681R/H mutations in SARS-CoV-2 VoCs Alpha, Delta, Omicron and viruses with D614G plus key signature mutations in spike protein alters fusogenicity and infectivity. Med Microbiol Immunol 212:103–122. https://doi.org/10.1007/s00430-022-00760-7
Kim Y, Jang G, Lee D et al (2022) Trypsin enhances SARS-CoV-2 infection by facilitating viral entry. Arch Virol 167:441–458. https://doi.org/10.1007/s00705-021-05343-0
Kumari A, Mittal L, Srivastava M et al (2021) Conformational characterization of the co-activator binding site revealed the mechanism to achieve the bioactive state of FXR. Front Mol Biosci 8:658312. https://doi.org/10.3389/fmolb.2021.658312
Kumari A, Mittal L, Srivastava M et al (2023) Deciphering the structural determinants critical in attaining the FXR partial agonism. J Phys Chem B 127:465–485. https://doi.org/10.1021/acs.jpcb.2c06325
Li F (2016) Structure, function, and evolution of coronavirus spike proteins. Annu Rev Virol 3:237–261. https://doi.org/10.1146/annurev-virology-110615-042301
Lin L, Li Q, Wang Y, Shi Y (2021) Syncytia formation during SARS-CoV-2 lung infection: a disastrous unity to eliminate lymphocytes. Cell Death Differ 28:2019–2021. https://doi.org/10.1038/s41418-021-00795-y
Lubinski B, Fernandes MHv, Frazier L et al (2022a) Functional evaluation of the P681H mutation on the proteolytic activation of the SARS-CoV-2 variant B.1.1.7 (Alpha) spike. iScience 25:103589. https://doi.org/10.1016/j.isci.2021.103589
Lubinski B, Frazier LE, Phan MVT et al (2022b) Spike protein cleavage-activation mediated by the SARS-CoV-2 P681R mutation: a case-study from its first appearance in variant of interest (VOI) A.23.1 identified in Uganda. bioRxiv. https://doi.org/10.1101/2021.06.30.450632
Lubinski B, Jaimes JA, Whittaker GR (2022c) Intrinsic furin-mediated cleavability of the spike S1/S2 site from SARS-CoV-2 variant B.1.1.529 (Omicron). bioRxiv. https://doi.org/10.1101/2022.04.20.488969
Maldonado LL, Bertelli AM, Kamenetzky L (2021) Molecular features similarities between SARS-CoV-2, SARS, MERS and key human genes could favour the viral infections and trigger collateral effects. Sci Rep 11:4108. https://doi.org/10.1038/s41598-021-83595-1
Matsuyama S, Ujike M, Morikawa S et al (2005) Protease-mediated enhancement of severe acute respiratory syndrome coronavirus infection. Proc Natl Acad Sci U S A 102:12543–12547. https://doi.org/10.1073/pnas.0503203102
Mittal L, Tonk RK, Awasthi A, Asthana S (2021) Targeting cryptic-orthosteric site of PD-L1 for inhibitor identification using structure-guided approach. Arch Biochem Biophys 713:109059. https://doi.org/10.1016/j.abb.2021.109059
Mlcochova P, Kemp SA, Dhar MS et al (2021) SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature 599:114–119. https://doi.org/10.1038/s41586-021-03944-y
Mykytyn AZ, Breugem TI, Riesebosch S et al (2021) SARS-CoV-2 entry into human airway organoids is serine protease-mediated and facilitated by the multibasic cleavage site. Elife. https://doi.org/10.7554/eLife.64508
Ou T, Mou H, Zhang L et al (2021) Hydroxychloroquine-mediated inhibition of SARS-CoV-2 entry is attenuated by TMPRSS2. PLoS Pathog 17:e1009212. https://doi.org/10.1371/journal.ppat.1009212
Panwar S, Kumari A, Kumar H et al (2022) Structure-based virtual screening, molecular dynamics simulation and in vitro evaluation to identify inhibitors against NAMPT. J Biomol Struct Dyn 40:10332–10344. https://doi.org/10.1080/07391102.2021.1943526
Peacock TP, Goldhill DH, Zhou J et al (2021) The furin cleavage site in the SARS-CoV-2 spike protein is required for transmission in ferrets. Nat Microbiol 6:899–909. https://doi.org/10.1038/s41564-021-00908-w
Pooja M, Reddy GJ, Hema K et al (2021) Unravelling high-affinity binding compounds towards transmembrane protease serine 2 enzyme in treating SARS-CoV-2 infection using molecular modelling and docking studies. Eur J Pharmacol 890:173688. https://doi.org/10.1016/j.ejphar.2020.173688
Rey FA, Lok S-M (2018) Common features of enveloped viruses and implications for immunogen design for next-generation vaccines. Cell 172:1319–1334. https://doi.org/10.1016/j.cell.2018.02.054
Samal S, Das S, Boliar S et al (2018) Cell surface ectodomain integrity of a subset of functional HIV-1 envelopes is dependent on a conserved hydrophilic domain containing region in their C-terminal tail. Retrovirology 15:50. https://doi.org/10.1186/s12977-018-0431-4
Sanda M, Morrison L, Goldman R (2021) N- and O-glycosylation of the SARS-CoV-2 spike protein. Anal Chem 93:2003–2009. https://doi.org/10.1021/acs.analchem.0c03173
Shajahan A, Supekar NT, Gleinich AS, Azadi P (2020) Deducing the N- and O-glycosylation profile of the spike protein of novel coronavirus SARS-CoV-2. Glycobiology 30:981–988. https://doi.org/10.1093/glycob/cwaa042
Singh R, Bhardwaj VK, Sharma J et al (2021) Identification of potential plant bioactive as SARS-CoV-2 Spike protein and human ACE2 fusion inhibitors. Comput Biol Med 136:104631. https://doi.org/10.1016/j.compbiomed.2021.104631
Suri C, Awasthi A, Asthana S (2022) Crystallographic landscape provides molecular insights into the modes of action of diverse ROR-γt modulators. Drug Discov Today 27:652–663. https://doi.org/10.1016/j.drudis.2021.11.022
Thomas P, Smart TG (2005) HEK293 cell line: a vehicle for the expression of recombinant proteins. J Pharmacol Toxicol Methods 51:187–200. https://doi.org/10.1016/j.vascn.2004.08.014
Vankadari N (2020) Structure of furin protease binding to SARS-CoV-2 spike glycoprotein and implications for potential targets and virulence. J Phys Chem Lett 11:6655–6663. https://doi.org/10.1021/acs.jpclett.0c01698
Vankadari N, Ketavarapu V, Mitnala S et al (2022) Structure of human TMPRSS2 in complex with SARS-CoV-2 spike glycoprotein and implications for potential therapeutics. J Phys Chem Lett 13:5324–5333. https://doi.org/10.1021/acs.jpclett.2c00967
Vardhan S, Sahoo SK (2022) Virtual screening by targeting proteolytic sites of furin and TMPRSS2 to propose potential compounds obstructing the entry of SARS-CoV-2 virus into human host cells. J Tradit Complement Med 12:6–15. https://doi.org/10.1016/j.jtcme.2021.04.001
Walls AC, Tortorici MA, Snijder J et al (2017) Tectonic conformational changes of a coronavirus spike glycoprotein promote membrane fusion. Proc Natl Acad Sci 114:11157–11162. https://doi.org/10.1073/pnas.1708727114
Walls AC, Xiong X, Park Y-J et al (2019) Unexpected receptor functional mimicry elucidates activation of coronavirus fusion. Cell 176:1026-1039.e15. https://doi.org/10.1016/j.cell.2018.12.028
Weissenhorn W, Dessen A, Calder LJ et al (1999) Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol 16:3–9. https://doi.org/10.1080/096876899294706
Wrobel AG, Benton DJ, Xu P et al (2020) SARS-CoV-2 and bat RaTG13 spike glycoprotein structures inform on virus evolution and furin-cleavage effects. Nat Struct Mol Biol 27:763–767. https://doi.org/10.1038/s41594-020-0468-7
Wu Zhang X, Leng Yap Y (2004) Structural similarity between HIV-1 gp41 and SARS-CoV S2 proteins suggests an analogous membrane fusion mechanism. THEOCHEM 677:73–76. https://doi.org/10.1016/j.theochem.2004.02.018
Yeung ML, Teng JLL, Jia L et al (2021) Soluble ACE2-mediated cell entry of SARS-CoV-2 via interaction with proteins related to the renin-angiotensin system. Cell 184:2212-2228.e12. https://doi.org/10.1016/j.cell.2021.02.053
Zeng C, Evans JP, King T et al (2022) SARS-CoV-2 spreads through cell-to-cell transmission. Proc Natl Acad Sci U S A. https://doi.org/10.1073/pnas.2111400119
Zheng Y, Zhou L-L, Su Y, Sun Q (2021) Cell fusion in the pathogenesis of COVID-19. Mil Med Res 8:68. https://doi.org/10.1186/s40779-021-00348-x
Acknowledgements
We sincerely thank Prof Pramod Kumar Garg, Executive Director, THSTI for full support and valuable input and guidance. We thank Prof. S Pöhlmann, Infection Biology Unit, Göttingen, Germany for providing ACE2-Fc plasmids as a kind gift. We thank Dr. B Graham (VRC/NIAID/NIH) for providing us with the spike construct (SARS-2-CoV S 2P). The following reagent was obtained through BEI Resources, NIAID, NIH: Human Embryonic Kidney Cells (HEK293T) Expressing Human Angiotensin-Converting Enzyme 2, HEK293T-hACE2 Cell Line, NR-52511.The TMPRSS2 plasmid was a gift from Roger Reeves (Addgene plasmid # 53887).
Funding
This work was supported by the Department of Biotechnology (DBT), Govt. of India and THSTI core funding.
Author information
Authors and Affiliations
Contributions
SS and RK conceived the project. RK, BL, GK, VM and A.G conducted most of the experiments. DS and SA performed the molecular dynamic simulation study. SS and RK analyzed the data. SS wrote the original manuscript; RK and BL edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Khatri, R., Lohiya, B., Kaur, G. et al. Understanding the role of conserved proline and serine residues in the SARS-CoV-2 spike cleavage sites in the virus entry, fusion, and infectivity. 3 Biotech 13, 323 (2023). https://doi.org/10.1007/s13205-023-03749-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03749-y