Abstract
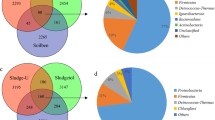
Toluene is one of the hydrocarbons that contaminate soil and groundwater, and has a high cost to remediate, which makes it an environmental pollutant of concern. This study aimed to find bacterial distribution from nonwoven geotextile (GT) fabric specimens in a pilot-scale permeable reactive barrier (PRB). Upon 167 days of incubation with the addition of toluene, the microbial community on the GT surfaces (n = 12) was investigated by the 16S rRNA metagenome sequencing approach. According to taxonomic classification, the Proteobacteria phylum dominated the metagenomes of all the geotextile samples (80–90%). Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway database search of the toluene degradation mechanism revealed the susceptible toluene-degrading species. For the toluene-to-benzoate degradation, the Cupriavidus genus, particularly C. gilardii, C. metallidurans, and C. taiwanensis, are likely to be functional. In addition to these species, the Novosphingobium genus was abundantly localized in the GTs, in particular Novosphingobium sp. ABRDHK2. The results suggested the biodegradation potential of these species in toluene remediation. Overall, this work sheds light on the variety of microorganisms found in the geotextile fabrics used in PRBs and the species involved in the biodegradation of toluene from several sources, including soil, sediment, and groundwater.






Similar content being viewed by others
Data availability
All data generated or analyzed during this study are included in this published article. The metagenome data are submitted to NCBI BioSample (SAMN28991445, SAMN28991445, SAMN28991446, SAMN28991447, SAMN28991448, SAMN28991449, SAMN28991450, SAMN28991451, SAMN28991452, SAMN28991453, and SAMN28991454) and BioProject (SUB11233613) databases.
References
Ali N, Dashti N, Khanafer M, Al-Awadhi H, Radwan S (2020) Bioremediation of soils saturated with spilled crude oil. Sci Rep 10(1):1–9
Balba M, Al-Awadhi N, Al-Daher R (1998) Bioremediation of oil-contaminated soil: microbiological methods for feasibility assessment and field evaluation. J Microbiol Methods 32:155–164
Beazley MJ, Martinez RJ, Rajan S, Powell J et al (2012) Microbial community analysis of a coastal salt marsh affected by the Deepwater Horizon oil spill. PLoS ONE 7(7):e41305
Benedek T, Szentgyörgyi F, Gergócs V, Menashe O et al (2021) Potential of Variovorax paradoxus isolate BFB1_13 for bioremediation of BTEX contaminated sites. AMB Express 11(1):1–17
BenIsrael M, Wanner P, Aravena R, Parker BL, Haack EA, Tsao DT, Dunfield KE (2019) Toluene biodegradation in the vadose zone of a poplar phytoremediation system identified using metagenomics and toluene-specific stable carbon isotope analysis. Int J Phytoremed 21(1):60–69
Bouhajja E, McGuire M, Liles MR, Bataille G, Agathos SN, George IF (2017) Identification of novel toluene monooxygenase genes in a hydrocarbon-polluted sediment using sequence-and function-based screening of metagenomic libraries. App Microbiol Biotech 101(2):797–808
Careghini A, Saponaro S, Sezenna E, Daghio M, Franzetti A, Gandolfi I, Bestetti G (2015) Lab-scale tests and numerical simulations for in situ treatment of polluted groundwater. J Hazard Mater 287:162–170
Chain PSG, Denef VJ, Konstantinidis KT, Vergez LM, Agulló L et al (2006) Burkholderia xenovorans LB400 harbors a multi-replicon, 9.73-Mbp genome shaped for versatility. Proc Natl Acad Sci USA 103:15280–15287
Choi DH, Kwon YM, Kwon KK, Kim SJ (2015) Complete genome sequence of Novosphingobium pentaromativorans US6-1(T). Stand Genomic Sci 10:107
Eze MO (2021) Metagenome analysis of a hydrocarbon-degrading bacterial consortium reveals the specific roles of BTEX biodegraders. Genes 12(1):98
Eze MO, Hose GC, George SC, Daniel R (2021) Diversity and metagenome analysis of a hydrocarbon-degrading bacterial consortium from asphalt lakes located in Wietze, Germany. AMB Express 11(1):1–12
Eze MO, Thiel V, Hose GC, George SC, Daniel R (2022a) Enhancing rhizoremediation of petroleum hydrocarbons through bioaugmentation with a plant growth-promoting bacterial consortium. Chemosphere 289:133143
Eze MO, Thiel V, Hose GC, George SC, Daniel R (2022b) Bacteria-plant interactions synergistically enhance biodegradation of diesel fuel hydrocarbons. Commun Earth Environ 3(1):1–10
Ghimire N, Kim B, Lee CM, Oh TJ (2022) Comparative genome analysis among Variovorax species and genome guided aromatic compound degradation analysis emphasizing 4-hydroxybenzoate degradation in Variovorax sp PAMC26660. BMC Genom 23(1):1–16
Gu M, Yin Q, Wu G (2021) Metagenomic analysis of facilitation mechanism for azo dye reactive red 2 degradation with the dosage of ferroferric oxide. J Water Process Eng 41:102010
Hashimoto T, Onda K, Morita T, Luxmy BS, Tada K, Miya A et al (2010) Contribution of the estrogen-degrading bacterium Novosphingobium sp strain JEM-1 to estrogen removal in wastewater treatment. J Environ Eng Asce 136:890–896
Hou Y, Zeng Q, Li H, Wu J, Xiang J, Huang H, Shi S (2022) Metagenomics-based interpretation of the impacts of silica nanoparticles exposure on phenol treatment performance in sequencing batch reactor system. Chem Eng J 428:132052
ITRC (2009) Evaluating natural source zone depletion at sites with LNAPL. The Interstate Technology & Regulatory Council, LNAPLs Team, 76
Janssen PJ, Van Houdt R, Moors H, Monsieurs P, Morin N, Michaux A, Benotmane MA, Leys N, Vallaeys T, Lapidus A, Monchy S, Medigue C, Taghavi S, McCorkle S, Dunn J, van der Lelie D, Mergeay M (2010) The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments. PLoS ONE 5(5):e10433
Jiang Y, Shao J, Wu X, Xu Y, Li R (2011) Active and silent members in the mlr gene cluster of a microcystin-degrading bacterium isolated from Lake Taihu, China. FEMS Microbiol Lett 322:108–114
Joshi MN, Dhebar SV, Dhebar SV, Bhargava P, Pandit A, Patel RP et al (2014) Metagenomics of petroleum muck: revealing microbial diversity and depicting microbial syntrophy. Arch Microbiol 196(8):531–544
Kalinovich I, Rutter A, Rowe RK, McWatters R, Poland JS (2008) The application of geotextile and granular filters for PCB remediation. Geosyn Int 15(3):173–183
Kim SJ, Park SJ, Cha IT, Min D, Kim JS, Chung WH et al (2014) Metabolic versatility of toluene-degrading, iron-reducing bacteria in tidal flat sediment, characterized by stable isotope probing-based metagenomic analysis. Environ Microbiol 16(1):189–204
Korkut EN, Martin JP, Yaman C (2006) Wastewater treatment with biomass attached to porous geotextile baffles. J Environ Eng 132:284–288
Lee Y, Jeon CO (2018) Paraburkholderia aromaticivorans sp. nov., an aromatic hydrocarbon-degrading bacterium, isolated from gasoline-contaminated soil. Int J Syst Evol Microbiol 68:1251–1257
Liu ZP, Wang BJ, Liu YH, Liu SJ (2005) Novosphingobium taihuense sp. nov, a novel aromatic-compound-degrading bacterium isolated from Taihu Lake, China. Int J Syst Evol Microbiol 55(3):1229–1232
Lovley DR (2003) Cleaning up with genomics: applying molecular biology to bioremediation. Nat Rev Microbiol 1(1):35–44
McWatters RS, Wilkins D, Spedding T, Hince G, Snape I, Rowe RK, et al (2014) Geosynthetics in barriers for hydrocarbon remediation in Antarctica. In: International Conference on Geosynthetics (pp1384–1391) Deutsche Gesellschaft fur Geotechnik eV
Mergeay M, Houba C, Gerits J (1978) Extrachromosomal inheritance controlling resistance to cadmium, cobalt, copper and zinc ions: evidence from curing in a Pseudomonas. Arch Int Physiol Biochim 86(2):440–442
Notomista E, Pennacchio F, Cafaro V, Smaldone G, Izzo V, Troncone L et al (2011) The marine isolate Novosphingobium sp PP1Y shows specific adaptation to use the aromatic fraction of fuels as the sole carbon and energy source. Microbial Ecol 61:582–594
Nwankwegu AS, Onwosi CO (2017) Bioremediation of gasoline contaminated agricultural soil by bioaugmentation. Environ Technol 7:1–11
Peng H, Zhang Q, Tan B, Li M, Zhang W, Feng J (2021) A metagenomic view of how different carbon sources enhance the aniline and simultaneous nitrogen removal capacities in the aniline degradation system. Bioresour Technol 335:125277
Pereira AC, Palakkeel Veetil D, Mulligan CN, Bhat S (2020) On-site nonwoven geotextile filtration method for remediation of lake water. In: CSCE Annual Conference
Polyak YM, Bakina LG, Chugunova MV, Mayachkina NV, Gerasimov AO, Bure VM (2018) Effect of remediation strategies on biological activity of oil-contaminated soil—a field study. Int Biodeterior Biodegradation 126:57–68
Posman KM, DeRito CM, Madsen EL (2017) Benzene degradation by a Variovorax species within a coal tar-contaminated groundwater microbial community. Appl Environ Microbiol 83(4):e02658-e2716
Rajamanickam R, Kaliyamoorthi K, Ramachandran N, Baskaran D, Krishnan J (2017) Batch biodegradation of toluene by mixed microbial consortia and its kinetics. Int Biodeterior Biodegr 119:282–288
Saxena R, Dhakan DB, Mittal P, Waiker P, Chowdhury A, Ghatak A, Sharma VK (2017) Metagenomic analysis of hot springs in Central India reveals hydrocarbon degrading thermophiles and pathways essential for survival in extreme environments. Front Microbiol 7:2123
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75(23):7537–7541
Schweitzer HD, Smith HJ, Barnhart EP, McKay LJ, Gerlach R, Cunningham AB et al (2022) Subsurface hydrocarbon degradation strategies in low-and high-sulfate coal seam communities identified with activity-based metagenomics. NPJ Biofilm Microbiom 8(1):1–10
Sheu SY, Huang CW, Chen JC, Chen ZH, Chen WM (2018) Novosphingobium arvoryzae spnov, isolated from a flooded rice field. Int J Syst Evol Microbiol 68:2151–2157
Silva CC, Hayden H, Sawbridge T, Mele P, Kruger RH, Rodrigues MV et al (2012) Phylogenetic and functional diversity of metagenomic libraries of phenol degrading sludge from petroleum refinery wastewater treatment system. AMB Express 2(1):1–13
Sohn JH, Kwon KK, Kang JH, Jung HB, Kim SJ (2004) Novosphingobium pentaromativorans spnov, a high-molecular-mass polycyclic aromatic hydrocarbon-degrading bacterium isolated from estuarine sediment. Int J System Evol Microbiol 54(5):1483–1487
Szentgyörgyi F, Benedek T, Fekete D, Táncsics A, Harkai P, Kriszt B (2022) Development of a bacterial consortium from Variovorax paradoxus and Pseudomonas veronii isolates applicable in the removal of BTEX. AMB Express 12(1):1–14
Tiirola MA, Mannisto MK, Puhakka JA, Kulomaa MS (2002) Isolation and characterization of Novosphingobium sp strain MT1, a dominant polychlorophenol-degrading strain in a groundwater bioremediation system. Appl Environ Microbiol 68:173–180
Wang J, Wang C, Li J, Bai P, Li Q, Shen M, et al (2018) Comparative genomics of degradative Novosphingobium strains with special reference to microcystin-degrading Novosphingobium spTHN1. Front Microbiol 2238
Wilhelm RC, Murphy SJ, Feriancek NM et al (2020) Paraburkholderia madseniana sp. nov., a phenolic acid-degrading bacterium isolated from acidic forest soil. Int J Syst Evol 70(3):2137–2146
Wood DE, Salzberg SL (2014) Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol 15(3):R46
Yadav R, Rajput V, Dharne M (2021) Functional metagenomic landscape of polluted river reveals potential genes involved in degradation of xenobiotic pollutants. Environ Res 192:110332
Yaman C (2020) Performance and kinetics of bioaugmentation, biostimulation, and natural attenuation processes for bioremediation of crude oil-contaminated soils. Processes 8(8):883
Yaman C, Martin JP, Korkut E (2005) Use of layered geotextiles to provide a substrate for biomass development in treatment of septic tank effluent prior to ground infiltration. J Environ Eng 131:1667–1675
Yaman C, Rehman S, Ahmad T, Kucukaga Y, Pala B, AlRushaid N, Riyaz Ul Hassan S, Yaman AB (2021) Community structure of bacteria and archaea associated with geotextile filters in anaerobic bioreactor landfills. Processes 9:1377
Yue JC, Clayton MK (2005) A similarity measure based on species proportions. Commun Stat-Theory Meth 34(11):2123–2131
Zhou L, Wang DW, Zhang SL et al (2020) Functional microorganisms involved in the sulfur and nitrogen metabolism in production water from a high-temperature offshore petroleum reservoir. Int Biodeterior Biodegr 154:105057
Funding
This research was funded by Imam Abdulrahman bin Faisal University (IAU) (Project No. 2019-037-Eng) through the Deanship of Scientific Research (DSR).
Author information
Authors and Affiliations
Contributions
CY conceptualized the study. CY, ABY, and IA setup the bioreactor system. IA, OA and STG analyzed the toluene content. EC conducted scanning electron microscopy. HT and AQ extracted the bacterial genome and worked on metagenome sequencing. HT and IB analyzed the metagenome data. HT, CY, OA, and IB wrote the manuscript. All authors read and commented on the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethics approval
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tombuloglu, H., Yaman, C., Boudellioua, I. et al. Metagenome analyses of microbial population in geotextile fabrics used in permeable reactor barriers for toluene biodegradation. 3 Biotech 13, 40 (2023). https://doi.org/10.1007/s13205-023-03460-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03460-y