Abstract
The development of artificial biocrust using cyanobacterium Phormidium tenue has been suggested as an effective strategy to prevent soil degradation. Here, a combination of in silico approaches with growth rate, photosynthetic pigment, morphology, and transcript analysis was used to identify specific genes and their protein products in response to 500 mM NaCl in P. tenue. The results show that 500 mM NaCl induces the expression of genes encoding glycerol-3-phosphate dehydrogenase (glpD) as a Flavoprotein, ribosomal protein S12 methylthiotransferase (rimO), and a hypothetical protein (sll0939). The constructed co-expression network revealed a group of abiotic stress-responsive genes. Using the Basic Local Alignment Search Tool (BLAST), the homologous proteins of rimO, glpD, and sll0939 were identified in the P. tenue genome. Encoded proteins of glpD, rimO, and DUF1622 genes, respectively, contain (DAO and DAO C), (UPF0004, Radical SAM and TRAM 2), and (DUF1622) domains. The predicted ligand included 22B and MG for DUF1622, FS5 for rimO, and FAD for glpD protein. There was no direct disruption in ligand-binding sites of these proteins by Na+, Cl−, or NaCl. The growth rate, photosynthetic pigment, and morphology of P. tenue were investigated, and the result showed an acceptable tolerance rate of this microorganism under salt stress. The quantitative real-time polymerase chain reaction (qRT-PCR) results revealed the up-regulation of glpD, rimO, and DUF1622 genes under salt stress. This is the first report on computational and experimental analyses of the glpD, rimO, and DUF1622 genes in P. tenue under salt stress to the best of our knowledge.










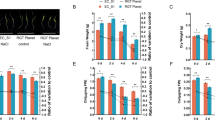
taken from three independent biological replicates. Different lower case letters above columns indicate significant differences at the (P < 0.05) and show significant differences compared with control
Similar content being viewed by others
References
Allakhverdiev SI, Murata N (2008) Salt stress inhibits photosystems II and I in cyanobacteria. Photosynth Res 98:529–539
Allakhverdiev SI, Sakamoto A, Nishiyama Y, Inaba M, Murata N (2000) Ionic and osmotic effects of NaCl-induced inactivation of photosystems I and II in Synechococcus sp. Plant Physiol 123:1047–1056
Allocco DJ, Kohane IS, Butte AJ (2004) Quantifying the relationship between co-expression, co-regulation and gene function. BMC Bioinform 5:1–10
Allu AD, Soja AM, Wu A, Szymanski J, Balazadeh S (2014) Salt stress and senescence: identification of cross-talk regulatory components. J Exp Bot 65:3993–4008
Antoninka A, Bowker MA, Reed SC, Doherty K (2016) Production of greenhouse-grown biocrust mosses and associated cyanobacteria to rehabilitate dryland soil function. Restor Ecol 24:324–335
Arakawa T, Timasheff S (1991) The interactions of proteins with salts, amino acids, and sugars at high concentration. In: Gilles R, Hoffmann EK, Bolis L (eds) Advances in Comparative and Environmental Physiology. Springer, Berlin, pp 226–245
Arora K, Kumar P, Bose D, Li X, Kulshrestha S (2021) Potential applications of algae in biochemical and bioenergy sector. 3 Biotech 11:1–24
Basak N, Krishnan V, Pandey V, Punjabi M, Hada A, Marathe A, Jolly M, Palaka BK, Ampasala DR, Sachdev A (2020) Expression profiling and in silico homology modeling of Inositol penta kis phosphate 2-kinase, a potential candidate gene for low phytate trait in soybean. 3 Biotech 10:1–21
Belnap J, Weber B, Büdel B (2016) Biological soil crusts as an organizing principle in drylands. In: Weber B, Büdel B, Belnap J (eds) Biological soil crusts: an organizing principle in drylands. Springer, Cham, pp 3–13
Bhardwaj AK, Shukla A, Mishra RK, Singh S, Mishra V, Uttam K, Singh MP, Sharma S, Gopal R (2017) Power and time dependent microwave assisted fabrication of silver nanoparticles decorated cotton (SNDC) fibers for bacterial decontamination. Front Microbiol 8:330
Bhardwaj AK, Shukla A, Maurya S, Singh SC, Uttam KN, Sundaram S, Singh MP, Gopal R (2018) Direct sunlight enabled photo-biochemical synthesis of silver nanoparticles and their Bactericidal Efficacy: photon energy as key for size and distribution control. J Photochem Photobiol B 188:42–49
Bhardwaj AK, Sundaram S, Yadav KK, Srivastav AL (2021) An overview of silver nano-particles as promising materials for water disinfection. Environ Technol Innov. https://doi.org/10.1016/j.eti.2021.101721
Borsani O, Valpuesta V, Botella MA (2001) Evidence for a role of salicylic acid in the oxidative damage generated by NaCl and osmotic stress in Arabidopsis seedlings. Plant Physiol 126:1024–1030
Bowker MA, Reed SC, Maestre FT, Eldridge DJ (2018) Biocrusts: the living skin of the earth. Springer
Bu C, Wang C, Yang Y, Zhang L, Bowker MA (2017) Physiological responses of artificial moss biocrusts to dehydration-rehydration process and heat stress on the Loess Plateau, China. J Arid Land 9:419–431
Büdel B, Dulić T, Darienko T, Rybalka N, Friedl T (2016) Cyanobacteria and algae of biological soil crusts. In: Weber B, Büdel B, Belnap J (eds) Biological soil crusts: an organizing principle in drylands. Springer, Cham, pp 55–80
Carpentier A-S, Torrésani B, Grossmann A, Hénaut A (2005) Decoding the nucleoid organisation of Bacillus subtilis and Escherichia coli through gene expression data. BMC Genomics 6:1–11
Chamizo S, Mugnai G, Rossi F, Certini G, De Philippis R (2018) Cyanobacteria inoculation improves soil stability and fertility on different textured soils: gaining insights for applicability in soil restoration. Front Environ Sci 6:49
Coleman RA, Lee DP (2004) Enzymes of triacylglycerol synthesis and their regulation. Prog Lipid Res 43:134–176
Colovos C, Yeates TO (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519
Consortium U (2019) UniProt: a worldwide hub of protein knowledge. Nucleic Acids Res 47:D506–D515
Cramer GR, Urano K, Delrot S, Pezzotti M, Shinozaki K (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol 11:1–14
Cruz de Carvalho R, dos Santos P, Branquinho C (2018) Production of moss-dominated biocrusts to enhance the stability and function of the margins of artificial water bodies. Restor Ecol 26:419–421
de Souza Silva CMM, Fay EF (2012) Effect of salinity on soil microorganisms. Soil Health Land Use Manage 10:177–198
Dickson DJ, Luterra MD, Ely RL (2012) Transcriptomic responses of Synechocystis sp. PCC 6803 encapsulated in silica gel. Appl Microbiol Biotechnol 96:183–196
Doi Y (2019) Glycerol metabolism and its regulation in lactic acid bacteria. Appl Microbiol Biotechnol 103:5079–5093
Doran JW, Parkin TB (1994) Defining and assessing soil quality. Defin Soil Qual Sustain Environ 35:1–21
Eisen MB, Spellman PT, Brown PO, Botstein D (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci 95:14863–14868
Eramian D, Eswar N, Shen MY, Sali A (2008) How well can the accuracy of comparative protein structure models be predicted? Protein Sci 17:1881–1893
Eungrasamee K, Incharoensakdi A, Lindblad P, Jantaro S (2020) Synechocystis sp. PCC 6803 overexpressing genes involved in CBB cycle and free fatty acid cycling enhances the significant levels of intracellular lipids and secreted free fatty acids. Sci Rep 10:1–13
Fageria N, Baligar V (2008) Ameliorating soil acidity of tropical Oxisols by liming for sustainable crop production. Adv Agron 99:345–399
Fasani E, Manara A, Martini F, Furini A, DalCorso G (2018) The potential of genetic engineering of plants for the remediation of soils contaminated with heavy metals. Plant Cell Environ 41:1201–1232
Forouhar F, Arragain S, Atta M, Gambarelli S, Mouesca J-M, Hussain M, Xiao R, Kieffer-Jaquinod S, Seetharaman J, Acton TB (2013) Two Fe-S clusters catalyze sulfur insertion by radical-SAM methylthiotransferases. Nat Chem Biol 9:333
Georg J, Rosana ARR, Chamot D, Migur A, Hess WR, Owttrim GW (2019) Inactivation of the RNA helicase CrhR impacts a specific subset of the transcriptome in the cyanobacterium Synechocystis sp. PCC 6803. RNA Biol 16:1205–1214
Gião MS, Keevil CW (2014) Listeria monocytogenes can form biofilms in tap water and enter into the viable but non-cultivable state. Microb Ecol 67:603–611
Goyal D, Yadav A, Prasad M, Singh TB, Shrivastav P, Ali A, Dantu PK, Mishra S (2020) Effect of heavy metals on plant growth: an overview. In: Naeem M, Ansari AA, Gill SS (eds) Contaminants in Agriculture. Springer, Cham, pp 79–101
Hagemann M (2011) Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35:87–123
Hernandez-Prieto MA, Futschik ME (2012) CyanoEXpress: a web database for exploration and visualisation of the integrated transcriptome of cyanobacterium Synechocystis sp PCC6803. Bioinformation 8:634
Hindré T, Knibbe C, Beslon G, Schneider D (2012) New insights into bacterial adaptation through in vivo and in silico experimental evolution. Nat Rev Microbiol 10:352–365
Hug JJ, Krug D, Müller R (2020) Bacteria as genetically programmable producers of bioactive natural products. Nat Rev Chem 4:172–193
Humphrey W, Dalke A, Schulten K (1996) VMD: visual molecular dynamics. J Mol Graph 14:33–38
Ishida TA, Nara K, Ma S, Takano T, Liu S (2009) Ectomycorrhizal fungal community in alkaline-saline soil in northeastern China. Mycorrhiza 19:329–335
Jarrett JT (2015) The biosynthesis of thiol-and thioether-containing cofactors and secondary metabolites catalyzed by radical S-adenosylmethionine enzymes. J Biol Chem 290:3972–3979
Jiang L, Pei H, Hu W, Ji Y, Han L, Ma G (2015) The feasibility of using complex wastewater from a monosodium glutamate factory to cultivate Spirulina subsalsa and accumulate biochemical composition. Biores Technol 180:304–310
Jugder BE, Ertan H, Wong YK, Braidy N, Manefield M, Marquis CP, Lee M (2016) Genomic, transcriptomic and proteomic analyses of Dehalobacter UNSWDHB in response to chloroform. Environ Microbiol Rep 8:814–824
Kakeh J, Gorji M, Mohammadi MH, Asadi H, Khormali F, Sohrabi M, Cerdà A (2020) Biological soil crusts determine soil properties and salt dynamics under arid climatic condition in Qara Qir. Science of The Total Environment, Iran, p 139168
Kakeh J, Gorji M, Mohammadi MH, Asadi H, Khormali F, Sohrabi M (2021) Effect of biocrusts on profile distribution of soil water content and salinity at different stages of evaporation. J Arid Environ 191:104514
Kamonchanock E, Aran I, Lindblad P, Saowarath J (2020) Synechocystis sp. PCC 6803 overexpressing genes involved in CBB cycle and free fatty acid cycling enhances the significant levels of intracellular lipids and secreted free fatty acids. Sci Rep. https://doi.org/10.1038/s41598-020-61100-4
Kanesaki Y, Suzuki I, Allakhverdiev SI, Mikami K, Murata N (2002) Salt stress and hyperosmotic stress regulate the expression of different sets of genes in Synechocystis sp. PCC 6803. Biochem Biophys Res Commun 290:339–348
Khalid E, Babiker E, Tinay AE (2003) Solubility and functional properties of sesame seed proteins as influenced by pH and/or salt concentration. Food Chem 82:361–366
Kharwar S, Bhattacharjee S, Mishra AK (2021) Bioinformatics analysis of enzymes involved in cysteine biosynthesis: first evidence for the formation of cysteine synthase complex in cyanobacteria. 3 Biotech 11:1–15
Khator K, Shekhawat G (2020) Nitric oxide mitigates salt-induced oxidative stress in Brassica juncea seedlings by regulating ROS metabolism and antioxidant defense system. 3 Biotech 10:1–12
Khodadadi F, Tohidfar M, Vahdati K, Dandekar AM, Leslie CA (2020) Functional analysis of walnut polyphenol oxidase gene (JrPPO1) in transgenic tobacco plants and PPO induction in response to walnut bacterial blight. Plant Pathol 69:756–764
Kirsch F, Klähn S, Hagemann M (2019) Salt-regulated accumulation of the compatible solutes sucrose and glucosylglycerol in cyanobacteria and its biotechnological potential. Front Microbiol 10:2139
Klähn S, Hagemann M (2011) Compatible solute biosynthesis in cyanobacteria. Environ Microbiol 13:551–562
Lan C-Y, Lin K-H, Chen C-L, Huang W-D, Chen C-C (2020) Comparisons of chlorophyll fluorescence and physiological characteristics of wheat seedlings influenced by iso-osmotic stresses from polyethylene glycol and sodium chloride. Agronomy 10:325
Langridge P, Fleury D (2011) Making the most of ‘omics’ for crop breeding. Trends Biotechnol 29:33–40
Lanyi JK (1974) Salt-dependent properties of proteins from extremely halophilic bacteria. Bacteriol Rev 38:272
Laskowski RA, Watson JD, Thornton JM (2005) ProFunc: a server for predicting protein function from 3D structure. Nucleic Acids Res 33:W89–W93
Lázaro R, Cantón Y, Solé-Benet A, Bevan J, Alexander R, Sancho L, Puigdefábregas J (2008) The influence of competition between lichen colonization and erosion on the evolution of soil surfaces in the Tabernas badlands (SE Spain) and its landscape effects. Geomorphology 102:252–266
Liang Y, Li D, Chen Y, Cheng J, Zhao G, Fahima T, Yan J (2020) Selenium mitigates salt-induced oxidative stress in durum wheat (Triticum durum Desf.) seedlings by modulating chlorophyll fluorescence, osmolyte accumulation, and antioxidant system. 3 Biotech 10:1–14
Liang Z, Zhi H, Fang Z, Zhang P (2021) Genetic engineering of yeast, filamentous fungi and bacteria for terpene production and applications in food industry. Food Res Int 147:110487
Los DA, Suzuki I, Zinchenko VV, Murata N (2008) Stress responses in Synechocystis: regulated genes and regulatory systems. Caister Academic Press, Norfolk, pp 117–157
Lu Y, Cheng L (2021) Computational analysis of LexA regulons in Proteus species. 3 Biotech 11:1–15
Lu CH, Yu CS, Lin YF, Chen JY. Predicting flavin and nicotinamide adenine dinucleotide-binding sites in proteins using the fragment transformation method. Biomed Res Int. 2015; 2015:402536
Lu S, Wang J, Chitsaz F, Derbyshire MK, Geer RC, Gonzales NR, Gwadz M, Hurwitz DI, Marchler GH, Song JS (2020) CDD/SPARCLE: the conserved domain database in 2020. Nucleic Acids Res 48:D265–D268
Lynn TM, Zhran M, Wang LF, Ge T, Yu SS, Kyaw EP, Latt ZK, Htwe TM (2021) Effect of land use on soil properties, microbial abundance and diversity of four different crop lands in central Myanmar. 3 Biotech 11:1–15
Macindoe G, Mavridis L, Venkatraman V, Devignes M-D, Ritchie DW (2010) HexServer: an FFT-based protein docking server powered by graphics processors. Nucleic Acids Res 38:W445–W449
Maestre FT, Sole R, Singh BK (2017) Microbial biotechnology as a tool to restore degraded drylands. Microb Biotechnol 10:1250–1253
Mahdavi S, Razeghi J, Pazhouhandeh M, Movafeghi A, Kosari-Nasab M, Kianianmomeni A (2020) Characterization of two predicted DASH-related proteins from the green alga Volvox carteri provides new insights into their light-mediated transcript regulation and DNA repair activity. Algal Res 52:102116
Trisilowati, Mallet DG. In silico experimental modeling of cancer treatment. ISRN Oncol. 2012;2012:828701
Mathpal P, Kumar U, Kumar A, Kumar S, Malik S, Kumar N, Dhaliwal H, Kumar S (2018) Identification, expression analysis, and molecular modeling of Iron-deficiency-specific clone 3 (Ids3)-like gene in hexaploid wheat. 3 Biotech 8:1–11
Mironov KS, Sinetova MA, Shumskaya M, Los DA (2019) Universal molecular triggers of stress responses in Cyanobacterium Synechocystis. Life 9:67
Mugnai G, Rossi F, Felde VJMNL, Colesie C, Büdel B, Peth S, Kaplan A, De Philippis R (2018) Development of the polysaccharidic matrix in biocrusts induced by a cyanobacterium inoculated in sand microcosms. Biol Fertil Soils 54:27–40
Mukherjee A, Bhowmick S, Yadav S, Rashid MM, Chouhan GK, Vaishya JK, Verma JP (2021) Re-vitalizing of endophytic microbes for soil health management and plant protection. 3 Biotech 11:1–17
Murata N, Takahashi S, Nishiyama Y, Allakhverdiev SI (2007) Photoinhibition of photosystem II under environmental stress. Biochimica et Biophysica (BBA) Acta Bioenergetics 1767:414–421
Nagarajan S, Nagarajan S (2009) Abiotic tolerance and crop improvement. In: Pareek A, Sopory SK, Bohnert HJ (eds) Abiotic stress adaptation in plants. Springer, Dordrecht, pp 1–11
Namwong S, Hiraga K, Takada K, Tsunemi M, Tanasupawat S, Oda K (2006) A halophilic serine proteinase from Halobacillus sp. SR5-3 isolated from fish sauce: purification and characterization. Biosci Biotechnol Biochem 70:1395–1401
Naraian R (2019) Mycodegradation of Lignocelluloses. Springer
Nawaz K, Hussain K, Majeed A, Khan F, Afghan S, Ali K (2010) Fatality of salt stress to plants: morphological, physiological and biochemical aspects. Afr J Biotechnol 9:34
Obalum S, Chibuike G, Peth S, Ouyang Y (2017) Soil organic matter as sole indicator of soil degradation. Environ Monit Assess 189:176
Oshone R, Ngom M, Chu F, Mansour S, Sy MO, Champion A, Tisa LS (2017) Genomic, transcriptomic, and proteomic approaches towards understanding the molecular mechanisms of salt tolerance in Frankia strains isolated from Casuarina trees. BMC Genomics 18:1–21
Osman KT (2018) Saline and sodic soils. In: Osman KT (ed) Management of soil problems. Springer, Cham, pp 255–298
Ozturk S, Aslim B (2010) Modification of exopolysaccharide composition and production by three cyanobacterial isolates under salt stress. Environ Sci Pollut Res 17:595–602
Parmar N, Singh KH, Sharma D, Singh L, Kumar P, Nanjundan J, Khan YJ, Chauhan DK, Thakur AK (2017) Genetic engineering strategies for biotic and abiotic stress tolerance and quality enhancement in horticultural crops: a comprehensive review. 3 Biotech 7:1–35
Pei H, Jiang L, Hou Q, Yu Z (2017) Toward facilitating microalgae cope with effluent from anaerobic digestion of kitchen waste: the art of agricultural phytohormones. Biotechnol Biofuels 10:1–18
Phatak SS, Stephan CC, Cavasotto CN (2009) High-throughput and in silico screenings in drug discovery. Expert Opin Drug Discov 4:947–959
Punjabi M, Bharadvaja N, Sachdev A, Krishnan V (2018) Molecular characterization, modeling, and docking analysis of late phytic acid biosynthesis pathway gene, inositol polyphosphate 6-/3-/5-kinase, a potential candidate for developing low phytate crops. 3 Biotech 8:1–20
Qadir M, Ghafoor A, Murtaza G (2001) Use of saline–sodic waters through phytoremediation of calcareous saline–sodic soils. Agric Water Manag 50:197–210
Reed CJ, Lewis H, Trejo E, Winston V, Evilia C (2013) Protein adaptations in archaeal extremophiles. Archaea. https://doi.org/10.1155/2013/373275
Rezaei QBF, Solouki A, Tohidfar M, Zare Mehrjerdi M, Izadi-Darbandi A, Vahdati K (2020) Agrobacterium-mediated transformation of Persian walnut using BADH gene for salt and drought tolerance. J Horticult Sci Biotechnol. https://doi.org/10.1080/14620316.2020.1812446
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47–e47
Rocha F, Esteban Lucas-Borja M, Pereira P, Muñoz-Rojas M (2020) Cyanobacteria as a nature-based biotechnological tool for restoring salt-affected soils. Agronomy 10:1321
Roncero-Ramos B, Román J, Gómez-Serrano C, Cantón Y, Acién F (2019) Production of a biocrust-cyanobacteria strain (Nostoc commune) for large-scale restoration of dryland soils. J Appl Phycol 31:2217–2230
Rychlik W (2007) OLIGO 7 primer analysis software. PCR Prim Des. https://doi.org/10.1007/978-1-59745-528-2_2
Sahoo S, Mahapatra SR, Das N, Parida BK, Rath S, Misra N, Suar M (2020) Functional elucidation of hypothetical proteins associated with lipid accumulation: Prioritizing genetic engineering targets for improved algal biofuel production. Algal Res 47:101887
Schrödinger L, DeLano W (2020) PyMOL. Retrieved from http://www.pymol.org/pymol
Shen MY, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524
Singh S, Shrivastava AK (2017) In silico characterization and transcriptomic analysis of nif family genes from Anabaena sp. PCC7120. Cell Biol Toxicol 33:467–482
Smith P, House JI, Bustamante M, Sobocká J, Harper R, Pan G, West PC, Clark JM, Adhya T, Rumpel C (2016) Global change pressures on soils from land use and management. Glob Change Biol 22:1008–1028
Sommer V, Karsten U, Glaser K (2020) Halophilic algal communities in biological soil crusts isolated from potash tailings pile areas. Front Ecol Evol 8:46
Steele DJ, Franklin DJ, Underwood GJ (2014) Protection of cells from salinity stress by extracellular polymeric substances in diatom biofilms. Biofouling 30:987–998
Tonk L, Bosch K, Visser PM, Huisman J (2007) Salt tolerance of the harmful cyanobacterium Microcystis aeruginosa. Aquat Microb Ecol 46:117–123
Uchiyama J, Asakura R, Kimura M, Moriyama A, Tahara H, Kobayashi Y, Kubo Y, Yoshihara T, Ohta H (2012) Slr0967 and Sll0939 induced by the SphR response regulator in Synechocystis sp PCC 6803 are essential for growth under acid stress conditions. Biochimica et Biophysica Acta (BBA) Bioenergetics 1817:1270–1276
Uchiyama J, Asakura R, Moriyama A, Kubo Y, Shibata Y, Yoshino Y, Tahara H, Matsuhashi A, Sato S, Nakamura Y (2014) Sll0939 is induced by Slr0967 in the cyanobacterium Synechocystis sp. PCC6803 and is essential for growth under various stress conditions. Plant Physiol Biochem 81:36–43
Vonshak A, Guy R, Guy M (1988) The response of the filamentous cyanobacterium Spirulina platensis to salt stress. Arch Microbiol 150:417–420
Wan X-F, VerBerkmoes NC, McCue LA, Stanek D, Connelly H, Hauser LJ, Wu L, Liu X, Yan T, Leaphart A (2004) Transcriptomic and proteomic characterization of the Fur modulon in the metal-reducing bacterium Shewanella oneidensis. J Bacteriol 186:8385–8400
West NE (1990) Structure and function of microphytic soil crusts in wildland ecosystems of arid to semi-arid regions. Advances in ecological research. Elsevier, pp 179–223
Wu Y, Rao B, Wu P, Liu Y, Li G, Li D (2013) Development of artificially induced biological soil crusts in fields and their effects on top soil. Plant Soil 370 (1–2):115–124
Xin Z, Yong S, Yang L, Rong-Liang J, Xin-Rong L (2015) Osmotic adjustment of soil biocrust mosses in response to desiccation stress. Pedosphere 25:459–467
Yadav S, Modi P, Dave A, Vijapura A, Patel D, Patel M (2020) Effect of Abiotic Stress on Crops, Sustainable Crop Production. IntechOpen
Yamazaki K, Ishimori M, Kajiya-Kanegae H, Takanashi H, Fujimoto M, Yoneda J-I, Yano K, Koshiba T, Tanaka R, Iwata H (2020) Effect of salt tolerance on biomass production in a large population of sorghum accessions. Breed Sci 70:167–175
Yang Y, Guo Y (2018) Unraveling salt stress signaling in plants. J Integr Plant Biol 60:796–804
Yang J, Roy A, Zhang Y (2012) BioLiP: a semi-manually curated database for biologically relevant ligand–protein interactions. Nucleic Acids Res 41:D1096–D1103
Yang J, Roy A, Zhang Y (2013) Protein–ligand binding site recognition using complementary binding-specific substructure comparison and sequence profile alignment. Bioinformatics 29:2588–2595
Yang J, Anishchenko I, Park H, Peng Z, Ovchinnikov S, Baker D (2020) Improved protein structure prediction using predicted interresidue orientations. Proc Natl Acad Sci 117:1496–1503
Yeh JI, Chinte U, Du S (2008) Structure of glycerol-3-phosphate dehydrogenase, an essential monotopic membrane enzyme involved in respiration and metabolism. Proc Natl Acad Sci 105:3280–3285
Zhao Y, Li X, Wang F, Zhao X, Gao Y, Zhao C, He L, Li Z, Xu J (2018) Glycerol-3-phosphate dehydrogenase (GPDH) gene family in Zea mays L: Identification, subcellular localization, and transcriptional responses to abiotic stresses. PloS one 13:e0200357
Funding
No funding to declare.
Author information
Authors and Affiliations
Corresponding author
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Shahbazi, M., Tohidfar, M. & Azimzadeh Irani, M. Identification of the key functional genes in salt-stress tolerance of Cyanobacterium Phormidium tenue using in silico analysis. 3 Biotech 11, 503 (2021). https://doi.org/10.1007/s13205-021-03050-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-03050-w