Abstract
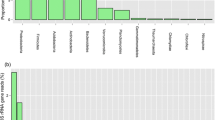
Intensive cropping degrades soil quality and disrupts the soil microbiome. To understand the effect of rice monocropping on soil-microbiome, we used a comparative 16S rRNA metagenome sequencing method to analyze the diversity of soil microflora at the genomic level. Soil samples were obtained from five locations viz., Chamarajnagara, Davangere, Gangavathi, Mandya, and Hassan of Karnataka, India. Chemical analysis of soil samples from these locations revealed significant variations in pH (6.00–8.38), electrical conductivity (0.17–0.69 dS m−1), organic carbon (0.51–1.29%), available nitrogen (279–551 kg ha−1), phosphorous (57–715 kg ha−1) and available potassium (121–564 kg ha−1). The 16S metagenome analysis revealed that the microbial diversity in Gangavathi soil samples was lower than in other locations. The soil sample of Gangavathi showed a higher abundance of Proteobacteria (85.78%) than Mandya (27.18%). The Firmicutes were more abundant in Chamarajnagar samples (36.01%). Furthermore, the KEGG pathway study revealed enriched nitrogen, phosphorus, and potassium metabolism pathways in all soil samples. In terms of the distribution of beneficial microflora, the decomposers were more predominant than the nutrient recyclers such as nitrogen fixers, phosphorous mineralizers, and nitrifiers. Furthermore, we isolated culturable soil microbes and tested their antagonistic activity in vitro against a fungal pathogen of rice, Magnaporthe oryzae strain MG01. Six Bacillus sp. and two strains of Trichoderma harzianum showed higher antagonistic activity against MG01. Our findings indicate that metagenome sequencing can be used to investigate the diversity, distribution, and abundance of soil microflora in rice-growing areas. The knowledge gathered can be used to develop strategies for maintaining soil quality and crop conservation to increase crop productivity.




Similar content being viewed by others
References
Ahmed I, Yokota A, Fujiwara T (2007) A novel highly boron tolerant bacterium, Bacillus boroniphilus sp. nov., isolated from soil, that requires boron for its growth. Extremophiles 11:217–224. https://doi.org/10.1007/s00792-006-0027-0
Anderson CR, Peterson ME, Frampton RA et al (2018) Rapid increases in soil pH solubilise organic matter, dramatically increase denitrification potential and strongly stimulate microorganisms from the Firmicutes phylum. PeerJ. https://doi.org/10.7717/peerj.6090
Arjun JK, Harikrishnan K (2011) Metagenomic analysis of bacterial diversity in the rice rhizosphere soil microbiome. Biotechnol Bioinf Bioeng 1(3):361–367
Bais HP, Weir TL, Perry LG et al (2006) The role of root exudates in rhizosphere interactions with plants and other organisms. Annu Rev Plant Biol 57:233–266. https://doi.org/10.1146/annurev.arplant.57.032905.105159
Baldrian P (2017) Forest microbiome: diversity, complexity and dynamics. FEMS Microbiol Rev 41:109–130. https://doi.org/10.1093/femsre/fuw040
Bano N, Hollibaugh JT (2000) Diversity and distribution of DNA sequences with affinity to ammonia-oxidizing bacteria of the beta subdivision of the class proteobacteria in the Arctic Ocean. Appl Environ Microbiol 66(5):1960–1969. https://doi.org/10.1128/aem.66.5.1960-1969.2000
Barnes RB, Richardson D, Berry JW, Hood RL (1945) Flame photometry a rapid analytical procedure. Indus Eng Chem Anal Edn 17(10):605–611
Barrow NJ (2017) The effects of pH on phosphate uptake from the soil. Plant Soil 410:401–410. https://doi.org/10.1007/s11104-016-3008-9
Bland JA, Brock TD (1973) The marine bacterium Leucothrix mucor as an algal epiphyte. Mar Biol 23:283–292. https://doi.org/10.1007/BF00389335
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Constancias F, Terrat S, Saby NPA et al (2015) Mapping and determinism of soil microbial community distribution across an agricultural landscape. Microbiologyopen 4:505–517. https://doi.org/10.1002/mbo3.255
Dai Z, Su W, Chen H et al (2018) Long-term nitrogen fertilization decreases bacterial diversity and favors the growth of Actinobacteria and Proteobacteria in agro-ecosystems across the globe. Glob Chang Biol 24:3452–3461. https://doi.org/10.1111/gcb.14163
Dickman SR, Bray RH (1940) Colorimetric determination of phosphate. Ind Eng Chem Anal Ed 12:665–668. https://doi.org/10.1021/ac50151a013
Ding LJ, Cui HL, Nie SA, Long XE, Duan GL, Zhu YG (2019) Microbiomes inhabiting rice roots and rhizosphere. FEMS Microbiol Ecol 95(5):fiz040. https://doi.org/10.1093/femsec/fiz040
Dubey A, Malla MA, Khan F et al (2019) Soil microbiome: a key player for conservation of soil health under changing climate. Biodivers Conserv 28:2405–2429. https://doi.org/10.1007/s10531-019-01760-5
Geisseler D, Scow KM (2014) Long-term effects of mineral fertilizers on soil microorganisms—a review. Soil Biol Biochem 75:54–63. https://doi.org/10.1016/j.soilbio.2014.03.023
Jackson M (1973) Soil chemical analysis. Prentice-Hall of India Private Limited, New Delhi, India, pp 498
Kraegeloh A, Amendt B, Kunte HJ (2005) Potassium transport in a halophilic member of the Bacteria domain: Identification and characterization of the K+ uptake systems TrkH and TrkI from Halomonas elongata DSM 2581T. J Bacteriol 187:1036–1043. https://doi.org/10.1128/JB.187.3.1036-1043.2005
Krige DG (1951) Journal of the chemical metallurgical & mining society of South Africa. J Chem Metall Soc South Min Africa 52:119–139
Liu M, Hu F, Chen X, Huang Q, Jiao J, Zhang B, Li H (2009) Organic amendments with reduced chemical fertilizer promote soil microbial development and nutrient availability in a subtropical paddy field: the influence of quantity, type and application time of organic amendments. Appl Soil Ecol 42(2):166–175
Long DH, Lee FN, TeBeest DO (2000) Effect of nitrogen fertilization on disease progress of rice blast on susceptible and resistant cultivars. Plant Dis 84:403–409. https://doi.org/10.1094/PDIS.2000.84.4.403
Nordlund S, Johansson M, Lindblad A, Norén A (1997) Nitrogen Fixation in Rhodospirillum rubrum: Regulation of Activity and Generation of Reductant. In: Biological Fixation of Nitrogen for Ecology and Sustainable Agriculture. Springer, pp 151–154
Ollivier J, Töwe S, Bannert A et al (2011) Nitrogen turnover in soil and global change. FEMS Microbiol Ecol 78:3–16. https://doi.org/10.1111/j.1574-6941.2011.01165.x
Oren A (2013) Life at High Salt Concentrations. In: Rosenberg E, DeLong EF, Lory S, Stackebrandt E, Thompson F (eds) The Prokaryotes. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-30123-0_57
Orr CH, James A, Leifert C et al (2011) Diversity and activity of free-living nitrogen-fixing bacteria and total bacteria in organic and conventionally managed soils. Appl Environ Microbiol 77:911–919. https://doi.org/10.1128/AEM.01250-10
Raaijmakers JM, Paulitz TC, Steinberg C, Alabouvette C, Moënne-Loccoz Y (2009) The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Pl Soil 321(1–2):341–361
Shen W, Lin X, Shi W, Min J, Gao N, Zhang H, Yin R, He X (2010) Higher rates of nitrogen fertilization decrease soil enzyme activities, microbial functional diversity and nitrification capacity in a Chinese polytunnel greenhouse vegetable land. Pl Soil 337(1–2):137–150
Song X, Tao B, Guo J et al (2018) Changes in the microbial community structure and soil chemical properties of vertisols under different cropping systems in northern China. Front Environ Sci 6:1–14. https://doi.org/10.3389/fenvs.2018.00132
Subbiah BV, Asija GL (1956) A rapid procedure for the estimation of available nitrogen in soils. Current Sci 25:259
Vincent JM (1947) Distortion of fungal hyphæ in the presence of certain inhibitors [20]. Nature 159:850. https://doi.org/10.1038/159850b0
Walkley A, Black IA (1934) An examination of the degtjareff method for determining soil organic matter, and a proposed modification of the chromic acid titration method. Soil Sci 37:29–38
Wieland G, Neumann R, Backhaus H (2001) Variation of microbial communities in soil, rhizosphere, and rhizoplane in response to crop species, soil type, and crop development. Appl Environ Microbiol 67:5849–5854. https://doi.org/10.1128/AEM.67.12.5849-5854.2001
Wu L, Li Z, Li J et al (2013) Assessment of shifts in microbial community structure and catabolic diversity in response to Rehmannia glutinosa monoculture. Appl Soil Ecol 67:1–9. https://doi.org/10.1016/j.apsoil.2013.02.008
Xuan DT, Guong VT, Rosling A, Alström S, Chai B, Högberg N (2012) Different crop rotation systems as drivers of change in soil bacterial community structure and yield of rice, Oryza sativa. Biol Fertil Soils 48(2):217–225
Yamamura S, Yamashita M, Fujimoto N et al (2007) Bacillus selenatarsenatis sp. nov., a selenate- and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int J Syst Evol Microbiol 57:1060–1064. https://doi.org/10.1099/ijs.0.64667-0
Zhalnina K, Dias R, de Quadros PD et al (2014) Soil pH determines microbial diversity and composition in the park grass experiment. Microb Ecol 69:395–406. https://doi.org/10.1007/s00248-014-0530-2
Zhang M, He Z (2004) Long-term changes in organic carbon and nutrients of an Ultisol under rice cropping in southeast China. Geoderma 118:167–179. https://doi.org/10.1016/S0016-7061(03)00191-5
Funding
This project was funded by Watershed Development Department, Government of Karnataka, India. Also, supported by Directorate of Research, University of Agricultural Sciences, Bangalore under Varietal Development Project ID. DR/VTDP/2017–18. The funding agency had no role in study design, data collection, analysis and interpretation, decision to publish, or preparation of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization- MKP, HBM; Data curation- AJD, BSR; Data analysis- LMN, BP, MEP, DP, AS; Fund acquisition- MKP; Project Supervision- MKP and HBM; Data Validation- MKP, HBM, DP, AJD and AS; Writing manuscript- LMP, HBM, MKP and AJD; Manuscript review and editing: HBM and MKP.
Corresponding author
Ethics declarations
Conflict of interests
The authors declare no conflict of interests.
Supplementary Information
Below is the link to the electronic supplementary material.



Rights and permissions
About this article
Cite this article
Prasannakumar, M.K., Netravathi, L.M., Mahesh, H.B. et al. Comparative metagenomic analysis of rice soil samples revealed the diverse microbial population and biocontrol organisms against plant pathogenic fungus Magnaporthe oryzae. 3 Biotech 11, 245 (2021). https://doi.org/10.1007/s13205-021-02783-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-021-02783-y