Abstract
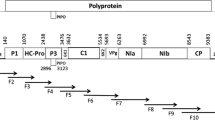
One of the destructive potyviruses which cause economic damage and serious yield losses to cucurbit crops around the world is Watermelon mosaic potyvirus. In 2016, 305 leaf samples from different cucurbit cultivars with deformation and reduction in leaf size, blistering, mild and severe mosaic symptoms were collected from different cucurbits-growing regions in Northwest of Iran. Total RNA and their cDNA were tested by RT-PCR assay using two sets of specific primers corresponding to the partial sequences of CP and P1 genomic regions, in which approximately 80 out of 305 samples were found to be infected by WMV. DNA fragments of about 780 bp and 545 bp in length were amplified that belonged to the CP and P1 genes, respectively. Phylogenetic trees of WMV isolates were clustered into three main independent groups with significant FST values (> 0.50 and > 0.55) for CP and P1 genes, respectively. dN/dS ratios obtained less than one (< 1) for CP gene that showed the WMV populations have been under the negative selection, whereas for P1 gene, the dN/dS values were calculated > 1 for EM clade containing; China, France, and Italy populations and < 1 for CL and G2 clades; South Korea and Iran populations. This results demonstrated that the WMV evolutionary selection pressure on the P1 gene is dependent on conditions such as the variety of cultivars and the type of cultivation.



Similar content being viewed by others
References
Ayazpour K, Vahidian M (2016) Study of watermelon mosaic virus in cucurbit fields of Jahrom area. Iran Vegetos 29:4. https://doi.org/10.5958/2229-4473.2016.00115.4
Bertin S, Manglli A, McLeish M, Tomassoli L (2020) Genetic variability of watermelon mosaic virus isolates infecting cucurbit crops in Italy. Adv Virol. https://doi.org/10.1007/s00705-020-04584-9
Chare ER, Holmes EC (2006) A phylogenetic survey of recombination frequency in plant RNA viruses. Adv Virol 151:933–946
Chung CT, Niemela SA, Miller RH (1989) One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci USA 86:2172–2175
Dombrovsky A, Huet H, Chejanovsky N, Raccah B (2005) Aphid transmission of a potyvirus depends on suitability of the helper component and the N terminus of the coat protein. Adv Virol 150:287–298
Desbiez C, Joannon B, Wipf-Scheibel C, Chandeysson C, Lecoq H (2011) Recombination in natural populations of watermelon mosaic virus: new agronomic threat or damp squib? J Gen Virol 92:1939–1948
Desbiez C, Lecoq H (2004) The nucleotide sequence of watermelon mosaic virus (WMV, Potyvirus) reveals interspecific recombination between two related potyviruses in the 5′ part of the genome. Adv Virol 149:1619–1632
Ebrahim-Nesbat F (1974) Distribution of watermelon mosaic viruses 1 and 2 in Iran. Phytopathology 79:352–358
Feinberg AP, Vogelstein B (1983) A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem 132:6–13
Fu YX, Li WH (1993) Statistical tests of neutrality of mutations. Genetics 133:693–709
Gadhave KR, Dutta B, Coolong T, Srinivasan R (2019) A non-persistent aphid-transmitted Potyvirus differentially alters the vector and non-vector biology through host plant quality manipulation. Sci Rep 9:1–12
Gao F, Lin W, Shen J, Liao F (2015) Genetic diversity and molecular evolution of arabis mosaic virus based on the CP gene sequence. Adv Virol 161:1047–1051
Garcia-Arenal F, Fraile A, Malpica JM (2003) Variation and evolution of plant virus populations. Int Microbiol 6:225–232
Glasa M, Bananej K, Predajňa L, Vahdat A (2011) Genetic diversity of watermelon mosaic virus in Slovakia and Iran shows distinct pattern. Plant Dis 95:38–42
Hajizadeh M, Bahrampour H, Abdollahzadeh J (2017) Genetic diversity and population structure of watermelon mosaic virus. J Plant Dis Prot 124:601–610
Hudson RR (2000) A new statistic for detecting genetic differentiation. Genetics 155:2011–2014
Abadkhah M, Koolivand D, Eini O (2018) A new distinct clade for iranian tomato spotted wilt virus isolates based on the polymerase, nucleocapsid, and non-structural genes. Plant Pathol J 34(6):514–531
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, Gibson TJ, Higgins DG (2007) Clustal W and Clustal X version 2.0. Bioinformatics 23(21):2947–2948
Lin CY, Ku HM, Chiang YH, Ho HY, Yu TA, Jan FJ (2012) Development of transgenic watermelon resistant to cucumber mosaic virus and watermelon mosaic virus by using a single chimeric transgene construct. Transgenic Res 21:983–993
Muhire BM, Varsani A, Martin DP (2014) SDT: a virus classification tool based on pairwise sequence alignment and identity calculation. PLoS ONE 9:e108277
Nei M, Tajima F (1981) DNA polymorphism detectable by restriction endonucleases. Genetics 97:145–163
Nematollahi S, Haghtaghi E, Koolivand D, Hajizadeh M (2014) Molecular detection of cucumber green mottle mosaic virus variants from cucurbits fields in Iran. Arch Phytopathol Plant Prot 47:1303–1310
Nigam D, LaTourrette K, Souza PFN, Garcia-Ruiz H (2019) Genome-wide variation in potyviruses. Front Plant Sci 10:1–28
Perotto MC, Pozzi EA, Celli MG, Luciani CE, Mitidieri MS, Conci VC (2017) Identifcation and characterization of a new potyvirus infecting cucurbits. Adv Virol. https://doi.org/10.1007/s00705-017-3660-2
Rajbanshi N, Ali A (2016) First complete genome sequence of a watermelon mosaic virus isolated from watermelon in the United States. Genome Announc 4(2):e00299-e316. https://doi.org/10.1128/genomeA.00299-16
Rozas J, Ferrer-Mata A, Sanchez-DelBarrio JC, Guirao-Rico S, Librado P, Ramos-Onsins SE, Sánchez-Gracia A (2017) DnaSP 6: DNA sequence polymorphism analysis of large datasets. Mol Biol Evol 34(12):3299–3302
Rubio L, Guerri J, Moreno P (2013) Genetic variability and evolutionary dynamics of viruses of the family Closteroviridae. Front Microbiol 4:1–15
Schneider WL, Roossinck MJ (2001) Genetic diversity in RNA virus ouasispecies is controlled by host–virus interactions. J Virol 75(14):6566–6571
Sharifi M, Massumi H, Heydarnejad J, Hosseini Pour A, Shaabanian M, Rahimian H (2008) Analysis of the biological and molecular variability of watermelon mosaic virus isolates from Iran. Virus Genes 37:304–313
Sokhandan-Bashir N, Melcher U (2012) Population genetic analysis of grapevine fanleaf virus. Adv Virol 157:1919–1929
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Tsompana M, Abad J, Purugganan M, Moyer JW (2005) The molecular population genetics of the tomato spotted wilt virus (TSWV) genome. Mol Ecol 14:53–66
Webb RE, Scott HA (1965) Isolation and identification of watermelon mosaic virus 1 and 2. Phytopathology 55:895–900
Acknowledgements
This research was supported by Islamic Azad University, Tabriz Branch, Tabriz-Iran. We would like to thanks from Miss Mahsa Abadkhah to valuable help in data analysis in this paper
Author information
Authors and Affiliations
Contributions
SN and DK conceived the project and designed the experiments; SN and NP performed the experiments; DK analyzed the data; DK wrote the paper; SN and DK revised the final version of the manuscript; all authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Rights and permissions
About this article
Cite this article
Nematollahi, S., Panahborhani, N. & Koolivand, D. Molecular characterization and population evolution analysis of Watermelon mosaic virus isolates on cucurbits of Northwest Iran. 3 Biotech 11, 43 (2021). https://doi.org/10.1007/s13205-020-02609-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02609-3