Abstract
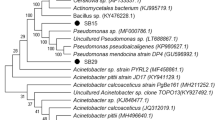
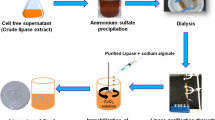
Organic solvent-tolerant lipase-producing microorganisms were isolated from petrol spilled soil. From ten morphologically distinct lipase-producing bacterial isolates, highest amount of lipase-producing isolate UBT1 was identified as Acinetobacter sp. using 16S rRNA gene sequencing (NCBI Accession No: MH879815). An increase in lipase production from 42 U/mL to 243 U/mL was obtained when different deoiled seed cakes were supplemented instead of olive oil in the medium. Further optimization of media components by the statistical approach assisted in discerning the main influencing media components and their optimum concentrations. Nine components glucose, castor seedcake, potassium nitrate, gum arabic, calcium chloride, magnesium sulphate, potassium di-hydrogen phosphate, dipotassium hydrogen phosphate, and ferric chloride were selected for Plackett–Burman design. The optimum concentrations of three significant selected components for the lipase production were found to be 0.025 gm% glucose, 0.002 gm% calcium chloride, and 0.2 gm% potassium di-hydrogen phosphate as determined by Response Surface Methodology. Increase in lipase production with 292.29 U/mL was achieved in the media containing optimized components and 2 gm% deoiled castor seed cake. Purification studies with ammonium sulphate precipitation, dialysis, and gel permeation chromatography resulted in 77.54% recovery with 5.77-fold partially purified lipase. The residual activity of lipase in 50 and 75% concentration of n-hexane among other solvents after 24 h was 105.05 and 90.42%, respectively, indicating its solvent tolerance. The present study reports the isolation of organic solvent-tolerant lipase-producing Acinetobacter sp. UBT1, optimization of the culture media for lipase production using the deoiled castor seed cake, and its partial purification.




Similar content being viewed by others
References
Ahmed EH, Raghavendra T, Madamwar D (2010). An alkaline lipase from organic solvent tolerant Acinetobacter sp. EH28: application for ethyl caprylate synthesis. Bioresour Technol, 101(10), 3628–3634. https://doi.org/https://doi.org/10.1016/j.biortech.2009.12.107
Allimoun MO, Ananzeh MR, Khleifat KM (2015). Screening selection and optimization of extracellular methanol and ethanol tolerant lipase from Acinetobacter sp. K5b4. Int J Biosci, 6(10), 44–56. http://dx.doi.org/https://doi.org/10.12692/ijb/6.10.44-56
Al-Limoun MO, Khleifat KM, Alsharafa KY, Qaralleh HN, Alrawashdeh SA (2019). Purification and characterization of a mesophilic organic solvent tolerant lipase produced by Acinetobacter sp. K5b4. Biocatal Biotranfor, 37(2), 139–151. https://doi.org/https://doi.org/10.1080/10242422.2018.1506445
Awad GE, Mostafa H, Danial EN, Abdelwahed NA, Awad HM (2015) Enhanced production of thermostable lipase from Bacillus cereus ASSCRC-P1 in waste frying oil based medium using statistical experimental design. J Appl Pharm Sci 5:7–15. https://doi.org/10.7324/JAPS.2015.50902
Bose A, Keharia H (2013) Production, characterization and applications of organic solvent tolerant lipase by Pseudomonas aeruginosa AAU2. Biocatal Agri Biotechnol 2(3):255–266. https://doi.org/10.1016/j.bcab.2013.03.009
Casas-Godoy L, Duquesne S, Bordes F, Sandoval G, Marty A (2012). Lipases: An overview. In Lipases and phospholipases (pp. 3–30). Humana Press. https://doi.org/https://doi.org/10.1007/978-1-61779-600-5_1
Chen SJ, Cheng CY, Chen TL (1998) Production of an alkaline lipase by Acinetobacter radioresistens. J Ferment Bioeng 86(3):308–312. https://doi.org/10.1016/S0922-338X(98)80135-9
Furini G, Berger JS, Campos JA, SAND ST, Germani JC, (2018) Production of lipolytic enzymes by bacteria isolated from biological effluent treatment systems. An Acad Bras Ciênc 90(3):2955–2965. https://doi.org/10.1590/0001-3765201820170952
Gonçalves Filho D, Silva AG, Guidini CZ (2019) Lipases: Sources, immobilization methods, and industrial applications. Appl Microbiol Biotechnol 103(18):7399–7423. https://doi.org/10.1007/s00253-019-10027-6
Gupta N, Sahai V, Gupta R (2007) Alkaline lipase from a novel strain Burkholderia multivorans: Statistical medium optimization and production in a bioreactor. Process Biochem 42(4):518–526. https://doi.org/10.1016/j.procbio.2006.10.006
Gururaj P, Ramalingam S, Devi GN, Gautam P (2016). Process optimization for production and purification of a thermostable, organic solvent tolerant lipase from Acinetobacter sp. AU07. Braz J Microbiol, 47(3), 647–657. https://doi.org/https://doi.org/10.1016/j.bjm.2015.04.002
Habibollahi H, Salehzadeh A (2018). Isolation, optimization, and molecular characterization of a lipase producing bacterium from oil contaminated soils. Pollut, 4(1), 119–128. https://doi.org/https://doi.org/10.22059/POLL.2017.238410.297
Haque E, Velmurugane J, Nagarajan J (2019). Media optimization for lipase production from Pseudomonas otitidis G5. J Adv Sci Res Manag, 4(7).
Hong MC, Chang MC (1998) Purification and characterization of an alkaline lipase from a newly isolated Acinetobacter radioresistens CMC-1. Biotechnol Lett 20(11):1027–1029. https://doi.org/10.1023/A:1005407005371
Ilesanmi OI, Adekunle AE, Omolaiye JA, Olorode EM, Ogunkanmi AL (2020) Isolation, optimization and molecular characterization of lipase producing bacteria from contaminated soil. Sci Afr 8:e00279
Jaeger KE, Reetz MT (1998) Microbial lipases form versatile tools for biotechnology. Trends Biotechnol 16(9):396–403. https://doi.org/10.1016/S0167-7799(98)01195-0
Jagtap S, Gore S, Yavankar S, Pardesi K, Chopade B (2010) Optimization of medium for lipase production by Acinetobacter haemolyticus from healthy human skin. Indian J Exp Biol 48(9):936–941
Jain R, Naik SN (2018) Adding value to the oil cake as a waste from oil processing industry: Production of lipase in solid state fermentation. Biocatal Agri Biotechnol 15:181–184. https://doi.org/10.1016/j.bcab.2018.06.010
Kapoor M, Gupta MN (2012) Lipase promiscuity and its biochemical applications. Process Biochem 47(4):555–569. https://doi.org/10.1016/j.procbio.2012.01.011
Khatape A, Chavan S, Khade S, Doiphode N (2015) Isolation and identification of lipid degrading microorganisms, optimization of medium and partial purification of the lipase enzyme. Int J Life Sci Res 3(1):99–103
Khoramnia A, Ebrahimpour A, Beh BK, Lai OM (2011). Production of a solvent, detergent, and thermotolerant lipase by a newly isolated Acinetobacter sp. in submerged and solid-state fermentations. J Biomed Biotechnol, 2011.
Kumar S, Kikon K, Upadhyay A, Kanwar SS, Gupta R (2005) Production, purification, and characterization of lipase from thermophilic and alkaliphilic Bacillus coagulans BTS-3. Protein Expr Purif 41(1):38–44. https://doi.org/10.1016/j.pep.2004.12.010
Kumar R, Mahajan S, Kumar A, Singh D (2011) Identification of variables and value optimization for optimum lipase production by Bacillus pumilus RK31 using statistical methodology. New Biotechnol 28(1):65–71. https://doi.org/10.1016/j.nbt.2010.06.007
Kumar A, Dhar K, Kanwar SS, Arora PK (2016) Lipase catalysis in organic solvents: advantages and applications. Biol Proced Online 18(1):2
Li J, Shen W, Fan G, Li X (2018). Screening, purification and characterization of lipase from Burkholderia pyrrocinia B1213. 3 Biotech, 8(9), 387. https://doi.org/https://doi.org/10.1007/s13205-018-1414-9
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ (1951) Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275
Masomian M, Rahman RNZRA, Salleh AB, Basri M (2010) A unique thermostable and organic solvent tolerant lipase from newly isolated Aneurinibacillus thermoaerophilus strain HZ: Physical factor studies. World J Microbiol Biotechnol 26(9):1693–1701. https://doi.org/10.1007/s11274-010-0347-1
Mohanasrinivasan V, Devi CS, Jayasmita D, Selvarajan E, Naine SJ (2018) Purification and characterization of extracellular lipase from Serratia marcescens VITSD2. Proc Natl Acad Sci India Sect B Biol Sci 88(1):373–381. https://doi.org/10.1007/s40011-016-0763-6
Noor I, Hasan M, Ramachandran K (2003) Effect of operating variables on the hydrolysis rate of palm oil by lipase. Process Biochem 39(1):13–20. https://doi.org/10.1016/S0032-9592(02)00263-7
Plackett RL, Burman JP (1946) The design of optimum multifactorial experiments. Biometrika 33(4):305–325. https://doi.org/10.2307/2332195
Prajapati V, Patel H, Trivedi U, Patel K (2014) Kinetic and thermodynamic characterization of lipase produced by Cellulomonas flavigena UNP3. J Basic Microbol 54(9):976–983. https://doi.org/10.1002/jobm.201300065
Ruchi G, Anshu G, Khare SK (2008) Lipase from solvent tolerant Pseudomonas aeruginosa strain: Production optimization by response surface methodology and application. Bioresour Technol 99(11):4796–4802. https://doi.org/10.1016/j.biortech.2007.09.053
Sadh PK, Duhan S, Duhan JS (2018) Agro-industrial wastes and their utilization using solid state fermentation: a review. Bioresour Bioprocess 5(1):1. https://doi.org/10.1186/s40643-017-0187-z
Sahoo RK, Kumar M, Mohanty S, Sawyer M, Rahman PK, Sukla LB, Subudhi E (2018) Statistical optimization for lipase production from solid waste of vegetable oil industry. Prep Biochem Biotechnol 48(4):321–326. https://doi.org/10.1080/10826068.2018.1431785
Sarac N, Ugur A (2016) A green alternative for oily wastewater treatment: lipase from Acinetobacter haemolyticus NS02-30. Desalin Water Treat 57(42):19750–19759. https://doi.org/10.1080/19443994.2015.1106346
Sarmah N, Revathi D, Sheelu G, Rani KY, Sridhar S, Mehtab V, Sumana C (2018) Recent advances on sources and industrial applications of lipases. Biotechnol Prog 34(1):5–28. https://doi.org/10.1002/btpr.2581
Sharma S (2014) Kanwar SS (2014). Organic solvent tolerant lipases and applications, The Sci World J
Sirisha E, Rajasekar N, Narasu ML (2010) Isolation and optimization of lipase producing bacteria from oil contaminated soils. Adv Biol Res 4(5):249–252
Soleymani S, Alizadeh H, Mohammadian H, Rabbani E, Moazen F, Sadeghi HM, Rabbani M (2017) Efficient media for high lipase production: One variable at a time approach. Avicenna J Med Biotechnol 9(2):82
Tembhurkar VR, Kulkarni MB, Peshwe SA (2012). Optimization of lipase production by Pseudomonas spp. in submerged batch process in shake flask culture. Sci Res Rep, 2(1), 46–50.
Veerapagu M, Narayanan AS, Ponmurugan K, Jeya KR (2013) Screening selection identification production and optimization of bacterial lipase from oil spilled soil. Asian J Pharm Clin Res 6(3):62–67
Winkler UK, Stuckmann MARTINA (1979) Glycogen, hyaluronate, and some other polysaccharides greatly enhance the formation of exolipase by Serratia marcescens. J Bacteriol 138(3):663–670
Zheng C (2018). Growth characteristics and enzyme production optimization of lipase producing strain. IOP Publishing. In IOP Conference Series: Earth and Environmental Science 108(4), 042087.
Acknowledgements
Accession number: The 16S rRNA of the Acinetobacter sp. UBT1 have been submitted to the NCBI gene bank under accession number MH879815.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Patel, R., Prajapati, V., Trivedi, U. et al. Optimization of organic solvent-tolerant lipase production by Acinetobacter sp. UBT1 using deoiled castor seed cake. 3 Biotech 10, 508 (2020). https://doi.org/10.1007/s13205-020-02501-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-020-02501-0