Abstract
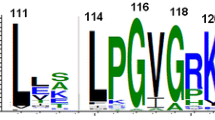
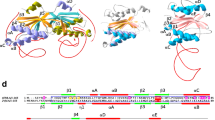
DNA repair is one of the key cellular events which balances between evolvability and integrity of the genome. Endonuclease IV enzymes are class II AP endonucleases under base excision repair pathway which act on abasic site and break the phosphodiester bond at the 5′ side. The role and activity of endonuclease IV proteins vary among different organisms; even it is absent in higher eukaryotes. The evolution of this protein family was studied by analyzing all homologs of the endonuclease IV protein family through different in silico techniques including phylogenetic tree generation and model building. The sequence analysis revealed four consensus sequence motifs within the AP2EC domain which are functionally important and conserved throughout the evolution process. It was also observed that the species and endonuclease IV gene evolution shape up differently in most of the organisms. Presence of the mitochondria-targeted signal peptides in fungal species Saccharomyces and Coccidioides suggest a possible endosymbiotic transfer of endonuclease IV genes to lower eukaryotes. Evolutionary changes among various clades in the protein-based phylogenetic tree have been investigated by comparison of homology models which suggests the conservation of overall fold of endonuclease IV proteins except for few alterations in loop orientation in few clades.







Similar content being viewed by others
References
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215(3):403–410
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Banner DW, Bloomer AC, Petsko GA, Phillips DC, Pogson CI, Wilson IA, Corran PH, Furth AJ, Milman JD, Offord RE, Priddle JD, Waley SG (1975) Structure of chicken muscle triose phosphate isomerase determined crystallographically at 2.5-angstrom resolution using amino acid sequence data. Nature 255(5510):609–614
Brochier C, Philippe H, Moreira D (2000) The evolutionary history of ribosomal protein RpS14: horizontal gene transfer at the heart of the ribosome. Trends Genet 16(12):529–533
Carstens BC, Knowles LL (2007) Estimating species phylogeny from gene-tree probabilities despite incomplete lineage sorting: an example form Melanoplus Grasshoppers. Syst Biol 56(3):400–411
Chan E, Weiss B (1987) Endonuclease IV of Escherichia coli is induced by paraquat. Proc Natl Acad Sci USA 84(10):3189–3193
Chatterjee N, Walker GC (2017) Mechanisms of DNA damage, repair, and mutagenesis. Environ Mol Mutagen 58(5):235–263
Cole JR, Wang Q, Cardenas E, Fish J, Chai B, Farris RJ, Kulam-Syed-Mohideen AS, McGarrell DM, Marsh T, Garrity GM, Tiedje JM (2009) The ribosomal database project: improved alignments and new tools for rRNA analysis. Nucleic Acids Res 37(Database issue):D141–D145
Crooks GE, Hon G, Chandonia JM, Brenner SE (2004) WebLogo: a sequence logo generator. Genome Res 14(6):1188–1190
Cunningham RP, Saporito SM, Spitzer SG, Weiss B (1986) Endonuclease IV (nfo) mutant of Escherichia coli. J Bacteriol 168(3):1120–1127
Daley JM, Zakaria C, Ramotar D (2010) The endonuclease IV family of apurinic/apyrimidinic endonucleases. Mutat Res 705:217–227
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39(4):783–791
Garrity GM, Holt JG (2001) The roadmap to the manual. In: Boone DR, Castenholz RW, Garrity GM (eds) Bergey’s manual of systematic bacteriology. The Archaea and the deeply branching and phototropic bacteria, vol 1, 2nd edn. Springer, New York, pp 119–166
Hosfield DJ, Guan Y, Haas BJ, Cunningham RP, Tainer JA (1999) Structure of the DNA repair enzyme endonuclease IV and its DNA complex: double-nucleotide flipping at abasic sites and three-metal-ion catalysis. Cell 98(3):397–408
Ide H, Tedzuka K, Shimzu H, Kimura Y, Purmal AA, Wallace SS, Kow YW (1994) Alpha-deoxyadenosine, a major anoxic radiolysis product of adenine in DNA, is a substrate for Escherichia coli endonuclease IV. Biochemistry 33(25):7842–7847
Jones DT, Taylor WR, Thornton JM (1992) The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci 8(3):275–282
Kanchan S, Mehrotra R, Chowdhury S (2015) In silico analysis of the endonuclease III protein family identifies key residues and processes during evolution. J Mol Evol 81(1–2):54–67
Marchler-Bauer A, Bryant SH (2004) CD-Search: protein domain annotations on the fly. Nucleic Acids Res 32(Web Server issue):W327–W331
Masson JY, Tremblay S, Ramotar D (1996) The Caenorhabditis elegans gene CeAPN1 encodes a homolog of Escherichia coli and yeast apurinic/apyrimidinic endonuclease. Gene 179(2):291–293
Mol CD, Hosfield DJ, Tainer JA (2000) Abasic site recognition by two apurinic/apyrimidinic endonuclease families in DNA base excision repair: the 3′ ends justify the means. Mutat Res 460(3–4):211–229
Moretti S, Armougom F, Wallace IM, Higgins DG, Jongeneel CV, Notredame C (2007) The M-Coffee web server: a meta-method for computing multiple sequence alignments by combining alternative alignment methods. Nucleic Acids Res 35(Web Server issue):W645–W648
Nei M, Rooney AP (2005) Concerted and birth-and-death evolution of multigene families. Annu Rev Genet 39:121–152
Puigbo P, Bravo IG, Garcia-Vallve S (2008) CAIcal: a combined set of tools to assess codon usage adaptation. Biol Direct 3:38
Puri RV, Singh N, Gupta RK, Tyagi AK (2013) Endonuclease IV is the major apurinic/apyrimidinic endonuclease in Mycobacterium tuberculosis and is important for protection against oxidative damage. PLoS ONE 8(8):e71535
Ramotar D, Popoff SC, Gralla EB, Demple B (1991) Cellular role of yeast Apn1 apurinic endonuclease/3′-diesterase: repair of oxidative and alkylation DNA damage and control of spontaneous mutation. Mol Cell Biol 11(9):4537–4544
Ramotar D, Vadnais J, Masson JY, Tremblay S (1998) Schizosaccharomyces pombe apn1 encodes a homologue of the Escherichia coli endonuclease IV family of DNA repair proteins. Biochim Biophys Acta 1396(1):15–20
Rice P, Longden I, Bleasby A (2000) EMBOSS: the European molecular biology open software suite. Trends Genet 16(6):276–277
Richardson CC, Kornberg A (1964) A deoxyribonucleic acid phosphatase-exonuclease from Escherichia coli. I. Purification of the enzyme and characterization of the phosphatase activity. J Biol Chem 239:242–250
Tamura K, Nei M (1993) Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol Biol Evol 10(3):512–526
Tamura K, Nei M, Kumar S (2004) Prospects for inferring very large phylogenies by using the neighbour-joining method. Proc Natl Acad Sci USA 101(30):11030–11035
Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S (2011) MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 28(10):2731–2739
Tsutakawa SE, Shin DS, Mol CD, Izumi T, Arvai AS, Mantha AK, Szczesny B, Ivanov IN, Hosfield DJ, Maiti B, Pique ME, Frankel KA, Hitomi K, Cunningham RP, Mitra S, Tainer JA (2013) Conserved structural chemistry for incision activity in structurally non-homologous apurinic/apyrimidinic endonuclease APE1 and endonuclease IV DNA repair enzymes. J Biol Chem 288(12):8445–8455
Whittaker RH (1969) New concepts of kingdoms or organisms. Evolutionary relations are better represented by new classifications than by the traditional two kingdoms. Science 163(3863):150–160
Zmasek CM, Eddy SR (2001) A simple algorithm to infer gene duplication and speciation events on a gene tree. Bioinformatics 17(9):821–828
Acknowledgements
The authors acknowledge the financial support from the University Grant Commission, Government of India in the form of Basic Science Research fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kanchan, S., Sharma, P. & Chowdhury, S. Evolution of endonuclease IV protein family: an in silico analysis. 3 Biotech 9, 168 (2019). https://doi.org/10.1007/s13205-019-1696-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1696-6