Abstract
Agrobacterium infection and regeneration of the putatively transformed plant from the explant remains arduous for some crop species like peanut. Henceforth, a competent and reproducible in planta genetic transformation protocol is established for peanut cv. CO7 by standardizing various factors such as pre-culture duration, acetosyringone concentration, duration of co-cultivation, sonication and vacuum infiltration. In the present investigation, Agrobacterium tumefaciens strain EHA105 harboring the binary vector pCAMBIA1301–bar was used for transformation. The two-stage selection was carried out using 4 and 250 mg l−1 BASTA® to completely eliminate the chimeric and non-transformed plants. The transgene integration into plant genome was evaluated by GUS histochemical assay, polymerase chain reaction (PCR), and Southern blot hybridization. Among the various combinations and concentrations analyzed, highest transformation efficiency was obtained when the 2-day pre-cultured explants were subjected to sonication for 6 min and vacuum infiltrated for 3 min in Agrobacterium suspension, and co-cultivated on MS medium supplemented with 150 µM acetosyringone for 3 days. The fidelity of the standardized in planta transformation method was assessed in five peanut cultivars and all the cultivars responded positively with a transformation efficiency ranging from minimum 31.3% (with cv. CO6) to maximum 38.6% (with cv. TMV7). The in planta transformation method optimized in this study could be beneficial to develop superior peanut cultivars with desirable genetic traits.





Similar content being viewed by others
Abbreviations
- CaMV 35S:
-
Cauliflower mosaic virus 35S promoter
- gusA :
-
β-Glucuronidase gene
- hptII:
-
Hygromycin phosphotransferase
- MS:
-
Murashige and Skoog medium
- nos Poly A :
-
Nopaline synthase terminator
References
Anuradha TS, Divya K, Jami SK, Kirti PB (2008) Transgenic tobacco and peanut plants expressing a mustard defensin show resistance to fungal pathogens. Plant Cell Rep 27:1777–1786
Athmaram TN, Bali G, Devaiah KM (2006) Integration and expression of Bluetongue VP2 gene in somatic embryos of peanut through particle bombardment method. Vaccine 24:2994–3000
Cervera M, Pina JA, Juarez J, Navarro L, Pena L (1998) Agrobacterium-mediated transformation of citrange: factors affecting transformation and regeneration. Plant Cell Rep 18:271–278
Chen X, Equi R, Baxter H, Berk K, Han J, Agarwal S, Zale J (2010) A high-throughput transient gene expression system for switchgrass seedlings. Biotechnol Biofuels 3:9
Chu Y, Bhattacharya A, Wu C, Knoll JE, Ozias-Akins P (2013) Improvement of peanut (Arachis hypogaea L.) transformation efficiency and determination of transgene copy number by relative quantitative real-time PCR. In Vitro Cell Dev Biol Plant 49:266–275
De Oliveira MLP, Febres VJ, Costa MGC, Moore GA, Otoni WC (2009) High-efficiency Agrobacterium-mediated transformation of citrus via sonication and vacuum infiltration. Plant Cell Rep 28:387–395
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA mini preparation: version II. Plant Mol Biol Rep 1:19–21
Egnin M, Mora A, Prakash CS (1998) Factors enhancing Agrobacterium tumefaciens-mediated gene transfer in peanut (Arachis hypogaea L.). In Vitro Cell Dev Biol Plant 34:310–318
Enserink M (2008) The peanut butter debate. Science 322:36–38
FAOSTAT (2014) Agricultural data. http://www.fao.org/faostat/en/#data/QC/visualize Accessed 16 Nov 2017
Feldmann KA, Marks MD (1987) Agrobacterium-mediated transformation of germinating seeds of Arabidopsis thaliana: a non-tissue culture approach. Mol Gen Genet 208:1–9
Fortin C, Nester EW, Dion P (1992) Growth inhibition and loss of virulence in cultures of Agrobacterium tumefaciens treated with acetosyringone. J Bacteriol 174:5676–5685
Franklin CI, Shorrosh KM, Trieu AN, Cassidy BG, Nelson RS (1993) Stable transformation of peanut callus via Agrobacterium-mediated DNA transfer. Transgenic Res 2:321–324
Haddadi F, Aziz MA, Abdullah SN, Tan SG, Kamaladini H (2015) An efficient Agrobacterium-mediated transformation of strawberry cv. Camarosa by a dual plasmid system. Molecules 20:3647–3666
Hood EE, Helmer GC, Fraley RT, Chilton MD (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in the region of pTiBo542 outside the T-DNA. J Bacteriol 168:1291–1301
Hood EE, Gelvin SB, Melchers LS, Hoekema A (1993) New Agrobacterium helper plasmids for gene transfer to plants. Transgen Res 2:208–218
Hsieh YF, Jain M, Jianping Wang J, Gallo M (2017) Direct organogenesis from cotyledonary node explants suitable for Agrobacterium-mediated transformation in peanut (Arachis hypogaea L.). Plant Cell Tissue Org Cult 128:161–175
Iqbal MM, Nazir F, Ali S, Asif MA, Zafar Y, Iqbal J, Ali GM (2012) Over expression of rice chitinase gene in transgenic peanut (Arachis hypogaea L.) improves resistance against leaf spot. Mol Biotechnol 50:129–136
Jaganath B, Subramanyam K, Mayavan S, Karthik S, Elayaraja D, Udayakumar R, Manickavasagam M, Ganapathi A (2014) An efficient in planta transformation of Jatropha curcas (L.) and multiplication of transformed plants through in vivo grafting. Protoplasma 251:591–601
Janila P, Nigam SN, Pandey MK, Nagesh P, Varshney RK (2013) Groundnut improvement: use of genetic and genomic tools. Front Plant Sci 4:23
Jefferson RA, Kavanagh TA, Bevan NW (1987) GUS fusions: β-glucuronidase as a sensitive and versatile gene fusion marker in higher plants. EMBO J 6:3901–3907
Krishna G, Singh BK, Kim EK, Morya VK, Ramteke PW (2015) Progress in genetic engineering of peanut (Arachis hypogaea L.)—a review. Plant Biotechnol J 13:147–162
Li S, Zhao DG, Wu YJ, Tian X (2009) A simplified seed transformation method for obtaining transgenic Brassica napus plants. Agric Sci China 8:658–663
Liu Z, Park BJ, Kanno A, Kameya T (2005) The novel use of a combination of sonication and vacuum infiltration in Agrobacterium-mediated transformation of kidney bean (Phaseolus vulgaris L.) with lea gene. Mol Breed 16:189–197
Manickavasagam M, Subramanyam K, Ishwarya R, Elayaraja D, Ganapathi A (2015) Assessment of factors influencing the tissue culture-independent Agrobacterium-mediated in planta genetic transformation of okra [Abelmoschus esculentus (L.) Moench]. Plant Cell Tissue Org Cult 123:309–320
Mariashibu TS, Subramanyam K, Arun M, Mayavan S, Rajesh M, Theboral J, Manickavasagam M, Ganapathi A (2013) Vacuum infiltration enhances the Agrobacterium-mediated genetic transformation in Indian soybean cultivars. Acta Physiol Plant 35:41–54
Mayavan S, Subramanyam K, Arun M, Rajesh M, Dev GK, Sivanandhan G, Jaganath B, Manickavasagam M, Selvaraj N, Ganapathi A (2013) Agrobacterium tumefaciens-mediated in planta seed transformation strategy in sugarcane. Plant Cell Rep 32:1557–1574
Mayavan S, Subramanyam K, Jaganath B, Sathish D, Manickavasagam M, Ganapthi A (2015) Agrobacterium-mediated in planta genetic transformation of sugarcane setts. Plant Cell Rep 34:1835–1848
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Park BJ, Liu Z, Kanno A, Kameya T (2005) Transformation of radish (Raphanus sativus L.) via sonication and vacuum infiltration of germinated seeds with Agrobacterium harbouring a group 3 LEA gene from B. napus. Plant Cell Rep 24:494–500
Pathak MR, Hamzah RY (2008) An effective method of sonicated assisted Agrobacterium-mediated transformation of chickpea. Plant Cell Tissue Org Cult 93:65–67
Qing CM, Fan L, Lei Y, Bouchez D, Tourneur C, Yan L, Robaglia C (2000) Transformation of Pakchoi (Brassica rapa L. ssp. chinensis) by Agrobacterium infiltration. Mol Breed 6:67–72
Qiusheng Z, Bao J, Likun L, Xianhua X (2005) Effects of antioxidants on the plant regeneration and gus expressive frequency of peanut (Arachis hypogaea) explants by Agrobacterium tumefaciens. Plant Cell Tissue Org Cult 81:83–90
Rohini VK, Rao KS (2000) Transformation of peanut (Arachis hypogaea L.): a non-tissue culture based approach for generating transgenic plants. Plant Sci 15:41–49
Rustom IYS, Lopez-Leiva MH, Nair BM (1996) Nutritional, sensory and physicochemical properties of peanut beverage sterilized under two different UHT conditions. Food Chem 56:45–53
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual, 2nd edn. Cold Spring Harbor Press, New York
Stachel SE, Messens E, Van Montagu M, Zambryski PC (1985) Identification of the signal molecules produced by wounded plant cells that activate T-DNA transfer in Agrobacterium tumefaciens. Nature 318:624–629
Subramaniam S, Samian R, Midrarullah Rathinam X (2009) Preliminary factors influencing transient expression of gus A in Dendrobium savin white protocorm-like bodies (PLBs) using Agrobacterium-mediated transformation system. WASJ 7:1295–1307
Subramanyam K, Subramanyam K, Sailaja KV, Srinivasulu M, Lakshmidevi K (2011) Highly efficient Agrobacterium-mediated transformation of banana cv. Rasthali (AAB) via sonication and vacuum infiltration. Plant Cell Rep 30:425–436
Subramanyam K, Rajesh M, Jaganath B, Vasuki A, Theboral J, Elayaraja D, Karthik S, Manickavasagam M, Ganapathi A (2013) Assessment of factors influencing the Agrobacterium-mediated in planta seed transformation of brinjal (Solanum melongena L.). Appl Biochem Biotechnol 171:45–468
Subramanyam K, Arunachalam C, Thaneswari RM, Sulaiman AA, Manickavasagam M, Ganapathi A (2015) Highly efficient Agrobacterium-mediated in planta genetic transformation of snake gourd (Tricosanthes cucumerina L.). Plant Cell Tissue Org Cult 123:133–142
Uranbey S, Sevimay CS, Kaya MD, Ipek A, Sancak C, Basalma D, Er C, Ozcan S (2005) Influence of different co-cultivation temperatures, periods and media on Agrobacterium tumefaciens-mediated gene transfer. Biol Plant 49:53–57
USDA (2016) Food composition database. https://ndb.nal.usda.gov/ndb/search/list?qlookup=16087 Accessed 16 Nov 2017
Yasmeen A, Mirza B, Inayatullah S, Safdar N, Jamil M, Ali S, Choudhry MF (2009) In planta transformation of tomato. Plant Mol Biol Rep 27:20–28
Acknowledgements
Sivabalan Karthik, is grateful to Jawaharlal Nehru Memorial Fund, New Delhi, India, for the award of Jawaharlal Nehru Scholarship (Ref no: SU-1/88/2016-17/79) for his doctoral research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13205_2018_1231_MOESM1_ESM.tif
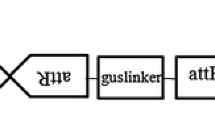
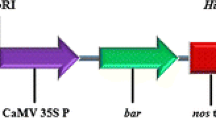
Supplementary material 1 (TIFF 76 kb) Supplementary Fig. 1 Illustration of the T-DNA region of the binary vector pCAMBIA1301–bar
13205_2018_1231_MOESM2_ESM.tif
Supplementary material 2 (TIFF 2296 kb) Supplementary Fig. 2 Evaluation of the germination percentage of 5 diverse peanut cultivars. One hundred seeds from each cultivar were used for the germination test. Mean values of three separate trials (±) with standard errors
Rights and permissions
About this article
Cite this article
Karthik, S., Pavan, G., Sathish, S. et al. Genotype-independent and enhanced in planta Agrobacterium tumefaciens-mediated genetic transformation of peanut [Arachis hypogaea (L.)]. 3 Biotech 8, 202 (2018). https://doi.org/10.1007/s13205-018-1231-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1231-1