Abstract
PCR-based fingerprinting using random amplified polymorphic DNA (RAPD) has been used widely for genome identification. In this study, 13 Salmonella Typhi strains were isolated from typhoid patients from Aswan, Cairo, Fayoum, and Monofya Governorates of Egypt. The isolates, along with three reference strains, i.e., O901, H901, and Ty2 were subjected to whole genome typing by RAPD PCR. Three RAPD-PCR 10-mer primers generated a total of 85 RAPD bands (81 polymorphic bands), 12 distinct PCR profiles, and proved to be useful for discriminating the isolates and strains studied. Interestingly, the B1 and C1 PCR profile were found only in Cairo and Monofya, respectively; and some PCR types appeared only in certain Governorates of Egypt. By combining the profiles obtained with the primer trio used in this study, an excellent discrimination index (D) of 0.942 was reached. Pairwise comparisons of Jaccard’s similarity coefficients calculated among the 12 PCR types identified three major clusters; i.e., O901 branch and Ty2 and H901 sub-branches. Principal component analysis adequately resolved each of these three major clusters. Three principal components accounted for about 72% of the variation, with the first two components accounting for about 62% of the total variance among the genotypes studied. Biclustering improved the display of groups of RAPD amplicons (markers) that cluster similarly across the genomes and could delineate features pertaining to genome structure. In conclusion, RAPD PCR provided a fast method with high potentials in surveillance and epidemiological investigations of Salmonella Typhi infections.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Typhoid fever is a systemic infection with the bacterium Salmonella enterica serotype Typhi. This fever is an important cause of illness and death with a global occurrence of 21.6 million infections and about 200,000 deaths from typhoid fever per year (Bhutta 2006). In Egypt, the population-based incidence of typhoid fever in Fayoum Governorate was 59/100,000 persons/year. A concerning prevalence of multidrug-resistant Salmonella Typhi (29%) was reported (Srinkantiah et al. 2006).
Salmonella Typhi has evolved a genetic mechanism for the expression of virulence genes located at pathogenicity islands in the bacterial genome. The size, structure, function, and distribution of these islands in the genomes of Salmonella subspecies and serovars can be markedly different (Hensel 2004; Aguirre et al. 2006; Akiba et al. 2006). As revealed by sequencing and microarray analyses, the genome of Salmonella Typhi also accumulated many pseudogenes (McClelland et al. 2004).
The success of epidemiological surveillance studies of Salmonella is related to the typing procedures applied to differentiate the genotypes. Several typing methods for Salmonella have been described for epidemiological and phylogenetic purposes (Ruiz et al. 2003; Lim et al. 2005; Araque 2009; Nath et al. 2010). The DNA-based typing methods are becoming increasingly useful for performing epidemiological surveys of pathogenic bacteria. Randomly amplified polymorphic DNA (RAPD) analysis, also known as arbitrarily primed-polymerase chain reaction (AP-PCR), is based on the presence of primer binding sites in the genome close enough to permit PCR amplification using a single primer with arbitrary nucleotide sequence at low annealing temperature. RAPD PCR, among other genetic typing procedures, has been shown to be useful tool to trace Salmonella epidemiologically and to distinguish Salmonella strains from different geographical origins. RAPD typing was useful for epidemiological typing of Salmonella isolates from human outbreaks and from avian sources and for complementing serotyping and phage typing methods (Soto et al. 1999, 2000; De Cesare et al. 2001; Lim et al. 2005; Quintaes et al. 2004; Smith et al. 2011).
The objective of this study is to develop a simple, easy to interpret, and low cost RAPD-based method, for typing Salmonella Typhi isolated from Egyptian patients suffering from typhoid fever. As RAPD PCR detects sequence diversity of total DNA, it is expected to display a good degree of sequence divergence. The method does not require any specific knowledge of the DNA sequence of the target organisms. This makes it a flexible tool that has great power and general applicability. Moreover, the method is more rapid and less technically demanding than most other molecular typing methods.
Materials and methods
Samples
Anti-coagulated whole peripheral blood (WPB) samples were collected from 300 patients suffering from prolonged fever and admitted to the Abbassia Fever Hospital (Cairo), Fayoum General Hospital (Fayoum), Aswan General Hospital (Aswan), and Shebein General Hospital (Monofya). Salmonella enterica serotype Typhi (Salmonella Typhi) was isolated by blood culture from 13 (4.3%) patients. Three reference Salmonella Typhi strains; i.e., TY2, O901, and H901 as well as E. coli JM109 strain (non-Salmonella control) were also included in this study (Table 1).
Materials
Bacterial identification kit was obtained from Bio-merieux Vitek Inc., (Hazelwood, Missouri, USA). Selective enrichment media (Selenite “F” enrichment broth) and plating media (MacConkey lactose bile salt agar and Salmonella–Shigella agar) were obtained from Becton, Dickinson Microbiology Systems (NJ, USA). Salmonella O and H antisera employed in the identification of somatic and flagellar antigens were obtained from SA Scientific Inc. (San Antonio, TX). Molecular biology grade reagents were purchased from Roche (Roche Diagnostics GmbH, Germany), New England Biolabs Inc. (MA, USA), FMC Bioproducts (Rockland, Maine, USA), and GeneCraft (GeneCraft GmbH, Germany).
Primers
Oligonucleotides were commercially synthesized by Operon (Operon, A Qiagen Company, Qiagen GmbH, Germany). Nine 10-mer primers were used for RAPD-PCR typing (Table 2). All primers used were resuspended in TE buffer, stored at −20 °C, and 10 μM (10 pm/μl) working solutions were prepared to be used in PCR.
Preparation of genomic DNA
Bacterial genomic DNA was prepared according to Sambrook et al. (2001). The bacteria were grown overnight at 37 °C in Luria–Bertani (LB) broth prior to DNA preparation. Lysozyme was used at a final concentration of 3 mg/ml. Sodium dodecyl sulfate (SDS) was added to final concentration of 1%, and proteinase K was used at a final concentration of 50 μg/ml. The prepared DNA was further purified using the High Pure PCR template preparation Kit (Roche Diagnostics GmbH, Germany) according to the protocol provided by the manufacturer. The quality of Salmonella Typhi DNA prepared in this study was verified by PCR amplification of the16S rDNA from the bacterial genome.
RAPD PCR
The reaction mixture (25 μl) contained 10 mM Tris–HCI pH 7.5, 50 mM KCI, 1.5 mM MgCl2, 0.5 mM spermidine, 0.1 mM dNTPs, 15 pmol of the RAPD primer, 20 ng genomic DNA, and 0.8 U of Taq DNA polymerase. Amplification was carried out in a heated-lid Biometra Thermal Cycler (Biometra GmbH, Germany) for 40 cycles, each consisting of a denaturing step of 1 min at 94 °C, followed by annealing step of 1 min at 36 °C and an extension step of 2 min at 72 °C. The last cycle was followed by 5 min of long extension at 72 °C. The amplification products were separated by gel electrophoresis in 2.2% agarose (SeaKem LE agarose; FMC Bioproducts, Rockland, Maine, USA) in 45 mM Tris–borate, 1 mM EDTA buffer (pH = 8.0), containing ethidium bromide at 0.5 μg/ ml at a constant voltage of 5 V/cm. The gels were photographed under UV transillumination using a digital camera inside a gel documentation system (Alpha Innotech, CA, USA).
Analysis of RAPD PCR Data
Gel images were analyzed for genetic similarity among isolates using the AlphaEase Software (Alpha Innotech, CA, USA). RAPD bands were scored as discrete variables, using “1” to indicate presence and “0” to indicate the absence of a band in the profile. The PCR profiles are defined by the pattern of presence or absence of bands on the gel. Each PCR profile was labeled with an alphabetical letter followed by a numeral subscript that identifies the primer used to generate the PCR profile. Each amplicon was assigned a name that begins with the letter P followed by a number that indicates the RAPD primer used, and a 2-digit number that identifies the band position. The discrimination index (D) was calculated for each primer by using Simpson’s index of diversity as described by Hunter and Gaston (1988) as follows:

Where N total number of strains in the population studied, S total number of RAPD types, n j number of strains belonging to the jth type, n j /N the probability that a single strain sampled at ransom will belong to the jth type, and n j (nj − 1)/N(N − 1) the probability that two strains sampled consecutively will belong to the same jth type.
The similarities between DNA fingerprints were calculated with the band-matching Jaccard’s coefficient that ranges from 0 to 1.0, where 1.0 represents 100% identity (presence and position) for all bands in the two PCR fingerprints being compared. A pairwise similarity (or distance) matrix was developed and cluster analysis was performed using the Unweighted Pair Group Method with Arithmetic averages (UPGMA) method. Principal component analysis (PCA), a mathematical procedure that uses orthogonal linear transformation, was used to recognize patterns in the RAPD-generated markers and to highlight the relationships between the genotypes examined. PCA and biclustering, i.e., clustering of RAPD types and RAPD amplicons were performed by using the Cluster and TreeView program of Stanford University, USA (Eisen et al. 1998).
Results and discussion
The Salmonella Typhi isolates as well as the three reference strains studied were found to give reproducible RAPD profiles from the same or from newly-extracted genomic DNA purified as described under “Materials and methods” and stored at −20 °C. The reproducibility of RAPD profiling, i.e., the ability to amplify the same RAPD profile and to observe the same polymorphism, was tested in at least three replicates.
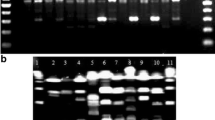
Out of the nine primers tested, only three primers, i.e., B02, E10, and E18 were suitable for discrimination (Table 2). Each of the three primers produced at least three distinct PCR profiles from the DNA preparations of Salmonella Typhi isolates and strains studied (Fig. 1), as well as, a distinct profile from E. coli (non-Salmonella control). The remaining primers produced less than 3 distinct PCR profiles. The PCR profiles generated by primers B02, E10, and E18 included 8–18 amplicons that ranged in size from 200–1,500 bp. These three primers were, therefore, used to reproducibly amplify random fragments of DNA from Salmonella Typhi genome.
Clustering of Salmonella Typhi RAPD profiles based on Jaccard’s coefficient and the UPGMA method. Band matching was based on the adjusted Rf values (AlphaEase FC Software, Alpha Innotech, CA, USA). The PCR profiles were labeled with alphabetical letters (A, B, C, and D) followed by the numerals (1, or 2, or 3) as subscripts. The numeral subscripts were respectively assigned to the three primers, B02, E10, and E18. a Profiles A1, B1a, B1b, C1, and D1 generated by RAPD primer B02. b Profiles A2, B2, and C2 generated by RAPD primer E10. c Profiles A3, B3, and C3 generated by RAPD primer E18. Reference profiles were A1, A2, and A3. Each amplicon was assigned a name that begins with the letter P followed by a number (1, or 2, or 3) that indicates the primer used (B02, E10, and E18, respectively), and a 2-digit number that identifies the band position. The direction of electrophoretic migration was from left to right, and band numbering starts from the large amplicons in each profile
A total of 85 RAPD bands (81 polymorphic bands) were scored. The four monomorphic bands were produced by RAPD primer E10 (P2-13 and P2-22) and primer E18 (P3-19 and P3-26). The numbers of RAPD bands resolved were 32 (all polymorphic, 39.5%), 25 (23 polymorphic, 28.4%), and 28 (26 polymorphic, 32.1%), for the primers B02, E10, and E18, respectively. On the average, 27 polymorphic RAPD markers were produced per RAPD primer from the studied genomes. The numbers of PCR profiles amplified by the three primers were 5, 3, and 3, respectively (Table 4).
The RAPD primer B02 showed the highest discriminatory index (D = 0.675) and identified five distinct PCR profiles (A1, B1a, B1b, C1, and D1) from 16 Salmonella Typhi samples examined (Fig. 1). The B1 and C1 PCR profiles were only found in the samples collected from Cairo and Fayoum, respectively (Table 3). This RAPD primer was useful for distinguishing the reference strain O901 (A1 PCR profile) from the rest of the Salmonella Typhi isolates and strains studied. The other two reference strains (Ty2 and H901) were also distinguishable by this primer and belonged to different PCR profiles (B1a and D1 profiles, respectively). Thus, the three reference strains yielded different band profiles with this primer. This observation agrees with those of McKenna et al. (1995) on the differentiation of Ty21a vaccine strain from the rest of Salmonella Typhi isolates using a single arbitrary primer, and those reported by Gürakan et al. (2008) on the use of a single RAPD primer as a serotype-specific marker for Salmonella Typhimurium.
The RAPD primer E10 had the lowest discriminatory index (D = 0.508) and identified only three distinct PCR profiles (A2, B2, and C2) from the isolates and strains examined. Two RAPD amplicons (P2-13 and P2-22) were common among these three PCR profiles (Fig. 1), while the other amplicons were useful for assigning Salmonella Typhi into the different profiles. The reference strains O901 and H901 had different PCR profiles with this primer. The RAPD primer E18 had an intermediate discriminatory index (D = 0.575) and identified three distinct PCR profiles (A3, B3, and C3) from the isolates and strains examined. Two RAPD amplicons (P3-19 and P3-26) were monomorphic among the three PCR profiles produced by this primer (Fig. 1). The reference strains H901 and Ty2 were indistinguishable by the two primers E10 or E18. Table (4) shows that the most frequent PCR profiles generated by primers B02, E10, and E18 were D1, B2, and C3, respectively. The D1 and B2 PCR profiles were found in three Governorates, while C3 PCR profile was found in the four Governorates.
In this study, the three RAPD primers used were able to generate 12 distinct PCR types by combining the profiles obtained with the three primers (Table 3). The most frequent PCR type was type 11 (4 isolates). A discrimination index (D) of 0.942 was achieved for the combined RAPD typing used in this study (Table 4). Lim et al. (2005) reported that a combination of two different RAPDs or a combination of RAPD and ERIC was better than the other combinations for the differentiation of field-isolated Salmonella strains and epidemiological studies. Combining the PCR types identified by the RAPD typing and phage typing or combining RAPD and PFGE increased the discrimination capacity (Soto et al. 1999; Delgado Ronda et al. 2006). The combination of RAPD typing with antibiotic susceptibility testing was a reliable discriminatory approach to differentiate Salmonella for epidemiologic purposes in Iran (Madadgar et al. 2008).
Jaccard’s similarity coefficients, based on the 85 RAPD markers, identified three major clusters, i.e., O901 branch and Ty2 and H901 sub-branches (Fig. 2a).
The O901 branch had the reference strain O901 (PCR type 1) along with PCR type 8. The Ty2 sub-branch contained the reference strain Ty2 along with five PCR types (i.e., Types 3–7), while the H901 sub-branch clustered the reference strain H901 (Type 9) along with three PCR types (Types 10–12). The reference strain H901 (Type 9) and the isolate A-Ash (Type 12) clustered closely in this sub-branch, with a Jaccard’s similarity coefficient of 0.82, indicating a close genetic similarity.
The results of principal component analysis (PCA), performed on the 85 RAPD markers, showed that the first (PC1), second (PC2), and third (PC3) principal components explained 48.52, 13.27, and 10.77% of the total variation in RAPD data, respectively. The first two components accounted for about 62% of the total variance, and the RAPD-PCR data were represented adequately by three principal components accounting for about 72% of the total variation. The results of PCA analysis were in considerable agreement with the overall representation of the genomes revealed by pairwise comparisons of Jaccard’s similarity coefficients. In the two-dimensional PCA plot, nearly all of the RAPD PCR types were clearly distinct from each other, and the major branch (O901 branch) and the two sub-branches (Ty2 and H901) could be resolved (Fig. 2b). Thus, the results of cluster analysis were supported by those of principal component analysis (PCA).
a The dendrogram constructed from the pairwise comparisons of Jaccard’s similarity coefficients and calculated based on the 85 RAPD markers. A major branch (O901 branch) was identified, as well as, two sub-branches (Ty2 and H901 sub-branches). b Results of principal component analysis (PCA) showing the two-dimensional (PC1 and PC2) plot. The first two principal components (PC1 and PC2) resolved the major branch (O901 branch) and each of the two sub-branches (Ty2 and H901). T1–T12 indicate the twelve RAPD PCR types
Apart from the most frequent PCR types (types 8 and 11), different PCR types were found in Cairo, Monofya, Fayoum, and Aswan (indicated by black-filled circles in Fig. 3). The most frequent PCR types (types 8 and 11) were not among the most similar PCR types (types 9 and 12). Likewise, the most frequent profiles generated by primers B02 and E10 were not among the most similar profiles generated by these primers (Table 4). The data, therefore, indicated that there is no close relationship between the most frequent PCR types observed. This supports the conclusion that the isolates of Salmonella Typhi had a considerable degree of genetic heterogeneity. However, due to the small sample size, the relationship between profiles and localities could not be established significantly. Previous studies on molecular typing and phylo-geographical distribution of Salmonella Typhi showed that different strains could be in circulation in endemic areas and outbreaks are related to only a few strains (Soto et al. 2000; Baker et al. 2010). Among Salmonella isolated in three provinces in the midwest of Spain, additional smaller clonal lines coexisted within every area. Most cases of infections are caused by the epidemic spread of a clone or, at least, by a group of strains that are genetically very close. Emergence and spread of Salmonella resulted mainly from the most frequent lineages (Soto et al. 2000; Delgado Ronda et al. 2006).
The biclustering algorithm was used to reorder the RAPD PCR types (genomes) and the RAPD PCR amplicons (markers) in order to optimize grouping and to visualize the data as a heat map. The biclustering indicated that certain RAPD amplicons (markers) cluster similarly across the PCR types (genomes). For example, RAPD amplicons P2-21 to P2-23 were absent from PCR types 1 and 8, while RAPD amplicons P2-07 to P2-08 were absent from PCR types 1, 8, and 12 (Fig. 4). On the other hand, RAPD amplicons P2-11 to P2-14 were only present in PCR types 1 and 8, while RAPD amplicons P2-03 to P2-24 were only present in PCR types 1, 8, and 12. RAPD amplicons P3-27 to P3-01 were present only in PCR types 2, 9, and 12.
Biclustering of RAPD PCR types (genomes) and RAPD PCR amplicons (markers). Each RAPD marker is represented by a single row and each PCR type is represented by a single column. The red color indicates the presence of the RAPD-PCR amplicon in the bacterial genome, while the black color indicates the absence of the amplicon in the bacterial genome. The two dendrograms were generated using the Cluster and TreeView program
It is conceivable that several features pertaining to genome architecture in Salmonella could be revealed by biclustering. The genome of Salmonella Typhi is characterized by the presence of a large number of genetic elements (Salmonella Pathogenicity Islands or SPI) that can be acquired by horizontal transmission to allow bacteria to rapidly gain complex virulence functions and to enable the emergence of new antibiotic-resistant epidemic strains. About 7.8% of the genome of Salmonella Typhi consists of pathogenicity islands and several common sequence motifs were identified between SPIs (Hensel 2004). These features, however, may be elucidated further using typing methods that use consensus oligonucleotides to reveal the presence of dispersed repetitive DNA sequences.
Due to the polymorphism inherent in the sequence, as well as, the short length of the RAPD primers used, the RAPD typing method resulted in a clustering of isolates into highly discriminating genetic trees. Although relatively few samples were used in this study, the data suggest that RAPD typing is discriminatory, it is easy to interpret and constitutes a low cost method to type the various strains of Salmonella Typhi. The high discriminatory power (D = 0.942) of the RAPD typing method used in this study revealed the robustness of the method and promises a high potential as a molecular typing method in surveillance and local outbreak investigations of Salmonella Typhi infections.
References
Aguirre A, Cabeza ML, Spinelli SV, McClelland M, Garcia Vescovi E, Soncini FC (2006) PhoP-induced genes within Salmonella pathogenicity island 1. J Bacteriol 188(19):6889–6898
Akiba M, Nakamura K, Shinoda D, Yoshii N, Ito H, Uchida I, Nakazawa M (2006) Detection and characterization of variant Salmonella genomic island 1 s from Salmonella Derby isolates. Jpn J Infect Dis. 59(5):341–345
Araque M (2009) Nontyphoid Salmonella gastroenteritis in pediatric patients from urban areas in the city of Merida, Venezuela. J Infect Dev Ctries 3(1):28–34
Baker S, Hanage WP, Holt KE (2010) Navigating the future of bacterial molecular epidemiology. Curr Opin Microbiol 13(5):640–645
Bhutta ZA (2006) Current concepts in the diagnosis and treatment of typhoid fever. BMJ 333:78–82
De Cesare A, Manfreda G, Dambaugh TR, Guerzoni ME, Franchini A (2001) Automated ribotyping and random amplified polymorphic DNA analysis for molecular typing of Salmonella enteritidis and Salmonella Typhimurium strains isolated in Italy. J Appl Microbiol 91:780–785
Delgado Ronda N, Muñoz Bellido JL, García García MI, Ibáñez Pérez R, Muñoz Criado S, Serrano Heranz R, Sáenz González MC, García Rodríguez JA (2006) Molecular epidemiology of drug-resistant Salmonella Typhimurium in Spain. Rev Esp Quimioterap 19(2):152–160
Eisen MB, Spellman PT, Brown PO, Botestein D (1998) Cluster analysis and display of genomic-wide expression patterns. Proc Natl Acad Sci USA 95:14863–14868
Gürakan GC, Aksoy C, Ögel ZB, Ören NG (2008) Differentiation of Salmonella Typhimurium from Salmonella Enteritidis and other Salmonella serotypes using random amplified polymorphic DNA analysis. Poult Sci 87:1068–1074
Hensel M (2004) Evolution of pathogenicity islands of Salmonellaenterica. Int J Med Microbiol 294(2–3):95–102
Hunter PR, Gaston MA (1988) Numerical index of the discriminatory ability of typing systems: an application of Simpson’s index of diversity. J Clin Microbiol 26:2465–2466
Lim H, Lee KH, Hong CH, Bahk GJ, Choi WS (2005) Comparison of four molecular typing methods for the differentiation of Salmonella spp. Int J Food Microbiol 105(3):411–418
Madadgar O, Tadjbakhsh H, Salehi TZ, Mahzounieh M, Feizabadi M (2008) Evaluation of random amplified polymorphic DNA analysis and antibiotic susceptibility application in discrimination of Salmonella Typhimurium isolates in Iran. New Microbiol 31:211–216
McClelland M, Sanderson KE, Clifton SW, Latreille P, Porwollik S, Sabo A, Meyer R, Bieri T, Ozersky P, McLellan M (2004) Comparison of genome degradation in ParaTyphi A and Typhi, human-restricted serovars of Salmonellaenterica that cause typhoid. Nat Genet 12:1268–1274
McKenna AJ, Bygraves JA, Maiden MC, Feavers IM (1995) Attenuated typhoid vaccine Salmonella Typhi Ty21a: fingerprinting and quality control. Microbiology 141:1993–2002
Nath G, Maurya P, Gulati AK (2010) ERIC PCR and RAPD based fingerprinting of Salmonella Typhi strains isolated over a period of two decades. Infect Genet Evol 10(4):530–536
Quintaes BR, Leal NC, Reis EM, Hofer E (2004) Optimization of randomly amplified polymorphic DNA-polymerase chain reaction for molecular typing of Salmonella enterica serovar Typhi. Rev Soc Bras Med Trop 37(2):143–147
Ruiz M, Rodriguez JC, Sirvent E, Escribano I, Cebrian L, Royo G (2003) Usefulness of different techniques in the study of the epidemiology of salmonellosis. APMIS 111(9):848–856
Sambrook J, MacCallum P, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, USA
Smith SI, Fowora MA, Goodluck HA, Nwaokorie FO, Aboaba OO, Opere B (2011) Molecular typing of Salmonella spp. isolated from food handlers and animals in Nigeria. Int J Mol Epidemiol Genet 2(1):73–77
Soto SM, Guerra B, González-Hevia MA, Mendoza MC (1999) Potential of three-way randomly amplified polymorphic DNA analysis as a typing method for twelve Salmonella serotypes. Appl Environ Microbiol 65(11):4830–4836
Soto SM, Martinez N, Guerra B, Gonzalez-Hevia MA, Mendoza MC (2000) Usefulness of genetic typing methods to trace epidemiologically Salmonella serotype Ohio. Epidemiol Infect 125:481–489
Srinkantiah P, Girgis F, Luby S, Jennings G, Wasfy M, Crump J, Hoekstra R, Anwer M, Mahoney F (2006) Population-based surveillance of typhoid fever in Egypt. Am J Trop Med Hyg 74(1):114–119
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rezk, N.A., Mansour, H., Ghoneim, N.H. et al. Typing of Salmonella Typhi strains isolated from Egypt by RAPD PCR. 3 Biotech 2, 17–25 (2012). https://doi.org/10.1007/s13205-011-0022-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13205-011-0022-8