Abstract
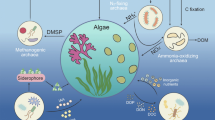
Ascidians host abundant and diverse bacterial symbionts, and ascidian-associated bacteria have been shown to benefit ascidian hosts. However, these host-symbiont interactions are often overlooked in coral reef ecosystems, where ascidians frequently occur as key components. In this study, we explored the bacterial communities associated with Atriolum robustum, a common ascidian from Xisha coral reef, China, using Illumina HiSeq high-throughput sequencing of the 16S rRNA gene amplicon. A total of 1411 OTUs were identified from A. robustum, revealing a high bacterial diversity with a Shannon index of 6.26 ± 1.04. Proteobacteria (in particular Alphaproteobacteria and Gammaproteobacteria) were dominant in A. robustum, followed by Bacteroidetes and Chloroflexi. Ten OTUs comprised the core microbiome of A. robustum, matching taxa with reported capabilities for heavy metal resistance, nutrient cycling, and defense against pathogens. Predictive metabolic profiles of the bacterial community associated with A. robustum, using Tax4Fun, showed a high abundance of genes involved in carbohydrate metabolism and amino acid metabolism. A comparison of the bacterial composition and predictive functions associated with A. robustum and other reported ascidians, suggested that horizontal transmission played a key role in the formation of bacterial communities in A. robustum. It was found that A. robustum from Xisha coral reef harbored a great diversity of bacteria, and bacterial symbionts had a potential functional role in metabolism and in the immune response of their ascidian host, and so played a beneficial role in maintaining the health of the coral reef ecosystem.






Similar content being viewed by others
References
Aßhauer KP, Wemheuer B, Daniel R, Meinicke P (2015) Tax4Fun: predicting functional profiles from metagenomic 16S rRNA data. Bioinformatics 31(17):2882–2884. https://doi.org/10.1093/bioinformatics/btv287
Banin E, Israely T, Kushmaro A, Loya Y, Orr E, Rosenberg E (2000) Penetration of the coral-bleaching bacterium Vibrio shiloi into Oculina patagonica. Appl Environ Microbiol 66(7):3031–3036. https://doi.org/10.1128/Aem.66.7.3031-3036.2000
Behrendt L, Larkum AWD, Trampe E, Norman A, Sorensen SJ, Kuhl M (2012) Microbial diversity of biofilm communities in microniches associated with the didemnid ascidian Lissoclinum patella. ISME J 6(6):1222–1237. https://doi.org/10.1038/ismej.2011.181
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R, Mills DA, Caporaso JG (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing[J]. Nat Methods 10(1):57–59. https://doi.org/10.1038/nmeth.2276
Bourne DG, Dennis PG, Uthicke S, Soo RM, Tyson GW, Webster N (2013) Coral reef invertebrate microbiomes correlate with the presence of photosymbionts. ISME J 7(7):1452–1458. https://doi.org/10.1038/ismej.2013.71
Buedenbender L, Carroll AR, Ekins M, Kurtboke DI (2017) Taxonomic and metabolite diversity of Actinomycetes associated with three Australian ascidians. Diversity-Basel 9(4):53. https://doi.org/10.3390/d9040053
Cahill PL, Fidler AE, Hopkins GA, Wood SA (2016) Geographically conserved microbiomes of four temperate water tunicates. Environ Microbiol Rep 8(4):470–478. https://doi.org/10.1111/1758-2229.12391
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Peña AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336. https://doi.org/10.1038/nmeth.f.303
Chen L, Fu CM, Wang GY (2017) Microbial diversity associated with ascidians: a review of research methods and application. Symbiosis 71(1):19–26. https://doi.org/10.1007/s13199-016-0398-7
Chen L, Hu JS, Xu JL, Shao CL, Wang GY (2018) Biological and chemical diversity of ascidian-associated microorganisms. Marine Drugs 16(10):362. https://doi.org/10.3390/md16100362
Dror H, Novak L, Evans JS, Lopez-Legentil S, Shenkar N (2019) Core and dynamic microbial communities of two invasive ascidians: can host-symbiont dynamics plasticity affect invasion capacity? Microb Ecol 78(1):170–184. https://doi.org/10.1007/s00248-018-1276-z
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998. https://doi.org/10.1038/Nmeth.2604
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Erwin PM, Pineda MC, Webster N, Turon X, Lopez-Legentil S (2013) Small core communities and high variability in bacteria associated with the introduced ascidian Styela plicata. Symbiosis 59(1):35–46. https://doi.org/10.1007/s13199-012-0204-0
Erwin PM, Pineda MC, Webster N, Turon X, Lopez-Legentil S (2014) Down under the tunic: bacterial biodiversity hotspots and widespread ammonia-oxidizing archaea in coral reef ascidians. ISME J 8(3):575–588. https://doi.org/10.1038/ismej.2013.188
Evans JS, Erwin PM, Shenkar N, Lopez-Legentil S (2017) Introduced ascidians harbor highly diverse and host-specific symbiotic microbial assemblages. Sci Rep 7:11033. https://doi.org/10.1038/s41598-017-11441-4
Evans JS, Lopez-Legentil S, Erwin PM (2018) Comparing two common DNA extraction kits for the characterization of symbiotic microbial communities from ascidian tissue. Microbes Environ 33(4):435–439. https://doi.org/10.1264/jsme2.ME18031
Farias P, Santo CE, Branco R, Francisco R, Santos S, Hansen L, Sorensen SJ, Morais P (2015) Natural hot spots for gain of multiple resistances: arsenic and antibiotic resistances in heterotrophic, aerobic bacteria from marine hydrothermal vent fields. Appl Environ Microbiol 81(7):2534–2543. https://doi.org/10.1128/Aem.03240-14
Hakim JA, Koo H, Kumar R, Lefkowitz EJ, Morrow CD, Powell ML, Watts AS, Bej AK (2016) The gut microbiome of the sea urchin, Lytechinus variegatus, from its natural habitat demonstrates selective attributes of microbial taxa and predictive metabolic profiles. FEMS Microbiol Ecol 92(9):32. https://doi.org/10.1093/femsec/fiw146
Hakim JA, Schram JB, Galloway AWE, Morrow CD, Crowley MR, Watts SA, Bej AK (2019) The purple sea urchin Strongylocentrotus purpuratus demonstrates a compartmentalization of gut bacterial microbiota, predictive functional attributes, and taxonomic co-occurrence. Microorganisms 7(2):35. https://doi.org/10.3390/microorganisms7020035
Hirose E (2015) Ascidian photosymbiosis: diversity of cyanobacterial transmission during embryogenesis. Genesis 53(1):121–131. https://doi.org/10.1002/dvg.22778
Huang H, Yang JH, Dong ZJ (2013) Coral reef atlas of Zhubi reef, Xisha Islands. China Ocean Press, Beijing
Huang H (2018) Coral reef atlas of Xisha Islands. Science Press, Beijing
Hussein ZAM, Bin Jantan I, Awaludin A, Ahmad NW (2002) Mosquito repellent activity of the methanol extracts of some ascidian species. Pharm Biol 40(5):358–361. https://doi.org/10.1076/phbi.40.5.358.8456
Hussein ZAM, Jantan I, Awaludin A, Ahmad NW (2001) Insecticidal activity of the methanol extracts of some tunicate species against Aedes aegypti and Anopheles maculatus. Pharm Biol 39(3):213–216. https://doi.org/10.1076/phbi.39.3.213.5922
Kehraus S, Gorzalka S, Hallmen C, Iqbal J, Muller CE, Wright AD, Wiese M, Konig GM (2004) Novel amino acid derived natural products from the ascidian Atriolum robustum: identification and pharmacological characterization of a unique adenosine derivative. J Med Chem 47(9):2243–2255. https://doi.org/10.1021/jm031092g
Krzmarzick MJ, Crary BB, Harding JJ, Oyerinde OO, Leri AC, Myneni SCB, Novak PJ (2012) Natural niche for organo halide-respiring chloroflexi. Appl Environ Microbiol 78(2):393–401. https://doi.org/10.1128/Aem.06510-11
Lopez-Legentil S, Turon X, Erwin PM (2016) Feeding cessation alters host morphology and bacterial communities in the ascidian Pseudodistoma crucigaster. Front Zool 13(1):2. https://doi.org/10.1186/s12983-016-0134-4
Magoc T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963. https://doi.org/10.1093/bioinformatics/btr507
Menezes CBA, Bonugli-Santos RC, Miqueletto PB, Passarini MRZ, Silva CHD, Justo MR, Leal RR, Fantinatti-Garboggini F, Oliveira VM, Berlinck RGS, Sette LD (2010) Microbial diversity associated with algae, ascidians and sponges from the north coast of Sao Paulo state, Brazil. Microbiol Res 165:466–482. https://doi.org/10.1016/j.micres.2009.09.005
Mitova M, Tommonaro G, Hentschel U, Muller WEG, De Rosa S (2004) Exocellular cyclic dipeptides from a Ruegeria strain associated with cell cultures of Suberites domuncula. Mar Biotechnol 6:95–103. https://doi.org/10.1007/s10126-003-0018-4
Miura N, Motone K, Takagi T, Aburaya S, Watanabe S, Aoki W, Ueda M (2019) Ruegeria sp. strains isolated from the reef-building coral Galaxea fascicularis inhibit growth of the temperature-dependent pathogen Vibrio coralliilyticus. Mar Biotechnol 21(1):1–8. https://doi.org/10.1007/s10126-018-9853-1
Moberg F, Folke C (1999) Ecological goods and services of coral reef ecosystems. Ecol Econ 29:215–233. https://doi.org/10.1016/S0921-8009(99)00009-9
Mohamed NM, Cicirelli EM, Kan JJ, Chen F, Fuqua C, Hill RT (2008) Diversity and quorum-sensing signal production of Proteobacteria associated with marine sponges. Environ Microbiol 10(1):75–86. https://doi.org/10.1111/j.1462-2920.2007.01431.x
Odum HT, Odum EP (1955) Trophic structure and productivity of a windward coral reef community on Eniwetok atoll. Ecol Monogr 2(3):291–320. https://doi.org/10.1016/j.ecolmodel.2003.12.004
Pandolfi JM, Bradbury RH, Sala E, Hughes TP, Bjorndal KA, Cooke RG, Mcardle D, Mcclenachan L, Newman MJH, Paredes G, Warner RR, Jackson JBC (2003) Global trajectories of the long-term decline of coral reef ecosystems. Science 301(5635):955–958. https://doi.org/10.1126/science.1085706
Pruesse E, Quast C, Knittel K, Fuchs BM, Ludwig WG, Peplies J, Glochner FO (2007) SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res 35(21):7188–7196. https://doi.org/10.1093/nar/gkm864
Ribes M, Coma R, Atkinson MJ, Kinzie RA (2005) Sponges and ascidians control removal of particulate organic nitrogen from coral reef water. Limnol Oceanogr 50(5):1480–1489. https://doi.org/10.4319/lo.2005.50.5.1480
Schreiber L, Kjeldsen KU, Funch P, Jensen J, Obst M, Lopez-Legentil S, Schramm A (2016) Endozoicomonas are specific, facultative symbionts of sea squirts. Front Microbiol 7:1042. https://doi.org/10.3389/fmicb.2016.01042
Shenkar N, Bronstein O, Loya Y (2008) Population dynamics of a coral reef ascidian in a deteriorating environment. Mar Ecol Prog Ser 367:163–171. https://doi.org/10.3354/meps07579
Stabili L, Licciano M, Longo C, Lezzi M, Giangrande A (2015) The Mediterranean non-indigenous ascidian Polyandrocarpa zorritensis: microbiological accumulation capability and environmental implications. Mar Pollut Bull 101(1):146–152. https://doi.org/10.1016/j.marpolbul.2015.11.005
Stevens JL, Jackson RL, Olson JB (2016) Bacteria associated with lionfish (Pterois volitans/miles complex) exhibit antibacterial activity against known fish pathogens. Mar Ecol Prog Ser 558:167–180. https://doi.org/10.3354/meps11789
Tebbett SB, Streit RP, Bellwood DR (2019) Expansion of a colonial ascidian following consecutive mass coral bleaching at Lizard Island, Australia. Mar Environ Res 144:125–129. https://doi.org/10.1016/j.marenvres.2019.01.007
Tianero MDB, Kwan JC, Wyche TP, Presson AP, Koch M, Barrows LR, Bugni TS, Schmidt EW (2015) Species specificity of symbiosis and secondary metabolism in ascidians. ISME J 9(3):615–628. https://doi.org/10.1038/ismej.2014.152
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261–5267. https://doi.org/10.1128/Aem.00062-07
Weil E, Smith G, Gil-Agudelo DL (2006) Status and progress in coral reef disease research. Dis Aquat Org 69(1):1–7. https://doi.org/10.3354/dao069001
Wilkinson C (2008) Status of coral reefs of the world: 2008. Global Coral Reef Monitoring Network and Reef and Rainforest Research Centre, Townsville
Yao QC, Yu KF, Liang JY, Wang YH, Hu BQ, Huang XY, Chen B, Qin ZJ (2019) The composition, diversity and predictive metabolic profiles of bacteria associated with the gut digesta of five sea urchins in Luhuitou fringing reef (northern South China Sea). Front Microbiol 10:e1168. https://doi.org/10.3389/fmicb.2019.01168
Zheng XQ, Li YC, Lin RC, Lan WL, Du JG, Yang XZ, Niu WT, Shi XF, Huang DY (2013) Coral reef conservation and restoration in mainland China. Malays J Sci 32:197–214 https://www.researchgate.net/publication/290950593
Funding
This research was funded by National Key R&D Program of China (2018YFD0900803), Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (Guangzhou) (GML2019ZD0605), Central Public-interest Scientific Institution Basal Research Fund, South China Sea Fisheries Research Institute, CAFS (NO. 2019TS28), Central Public-interest Scientific Institution Basal Research Fund, CAFS (NO. 2020TD16), Financial Fund of the Ministry of Agriculture and Rural Affairs, P. R. of China (NFZX2018).
Author information
Authors and Affiliations
Contributions
YL, PW, and CL conceived the research. YL, PW and YX performed the experiments. YL and PW wrote the manuscript. TW and LL contributed to sampling or data analysis. All authors edited and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 261 kb)
Rights and permissions
About this article
Cite this article
Liu, Y., Wu, P., Li, C. et al. The bacterial composition associated with Atriolum robustum, a common ascidian from Xisha coral reef, China. Symbiosis 83, 153–161 (2021). https://doi.org/10.1007/s13199-020-00742-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-020-00742-4