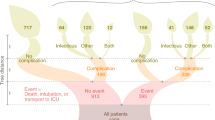
Abstract
The emergence of COVID-19 in late 2019 in Wuhan, China, has led to a global health crisis that has claimed many lives worldwide. A thorough understanding of the available COVID-19 datasets can enable healthcare professionals to identify cases at an early stage. This study presents an innovative pipeline-based framework for predicting survival and mortality in patients with COVID-19 by leveraging the Mexican COVID-19 patient dataset (COVID-19-MPD dataset). Preprocessing plays a pivotal role in ensuring that the framework delivers high-quality outcomes. We deploy various machine learning models with optimized hyperparameters within the framework. Through consistent experimental conditions and dataset utilization, we conducted multiple experiments employing diverse preprocessing techniques and models to maximize the area under the receiver operating characteristic curve (AUC) for COVID-19 prediction. Given the considerable dimensions of the dataset, feature selection is crucial for identifying factors influencing COVID-19 mortality or survival. We employ feature dimension reduction methods, such as principal component analysis and independent component analysis, in addition to feature selection techniques such as maximum relevance minimum redundancy and permutation feature importance. Impactful features related to patient outcomes can significantly aid experts in disease management by enhancing treatment efficacy and control measures. Following various experiments with standardized data and AUC assessment using the k-nearest neighbor algorithm with four components, the proposed framework achieves optimal results, attaining an AUC of 100%. Given its effectiveness in COVID-19 prediction, this framework has the potential for integration into medical decision support systems.
Graphical abstract














Similar content being viewed by others
Data availability
Data were used from a publicly available dataset [Online] Available: https://www.gob.mx/salud/documentos/datos-abiertos-152127
References
Abbas NAM, Salman HM (2020) Enhancing linear independent component analysis: comparison of various metaheuristic methods. Iraqi J Electr Electron Eng 16(1)
Abdulkareem NM, Abdulazeez AM, Zeebaree DQ, Hasan DA (2021) COVID-19 world vaccination progress using machine learning classification algorithms. Qubahan Acad J 1(2):100–105
Abnoosian K, Farnoosh R, Behzadi MH (2023a) A pipeline-based framework for early prediction of diabetes. J Health Biomed Inform 10(2):125–140
Abnoosian K, Farnoosh R, Behzadi MH (2023b) Prediction of diabetes disease using an ensemble of machine learning multiclassifier models. BMC Bioinformatics 24(1):337
Aguirre AA, Catherina R, Frye H, Shelley L (2020) Illicit wildlife trade, wet markets, and COVID-19: preventing future pandemics. World Medical & Health Policy 12(3):256–265
Akila A, Parameswari R, Jayakumari C (2022) Big data in healthcare: management, analysis, and future prospects. Handbook of Intelligent Healthcare Analytics: Knowledge Engineering with Big Data Analytics. https://doi.org/10.1002/9781119792550.ch14
Alkady W, ElBahnasy K, Leiva V, Gad W (2022) Classifying COVID-19 based on amino acids encoding with machine learning algorithms. Chemom Intell Lab Syst 224:104535
Althouse LA, Ware WB, Ferron JM (1998) Detecting departures from normality: a monte carlo simulation of a new omnibus test based on moments.
Bakar NA, Rosbi S (2020) Effect of coronavirus disease (COVID-19) to tourism industry. Int J Adv Eng Res Sci 7(4):189–193
Barut Z, Altuntaş V (2023) Comparison of performance of different k values with k-fold cross validation in a graph-based learning model for incrna-disease prediction. Kırklareli Üniversitesi Mühendislik Ve Fen Bilimleri Dergisi 9(1):63–82
Charbuty B, Abdulazeez A (2021) Classification based on decision tree algorithm for machine learning. J Appl Sci Technol Trends 2(01):20–28
Choo SW et al (2020) Are pangolins scapegoats of the COVID-19 outbreak-CoV transmission and pathology evidence? Conserv Lett 13(6):e12754
Claesen M, Simm J, Popovic D, Moreau Y, De Moor B (2014) Easy hyperparameter search using optunity. arXiv preprint arXiv:1412.1114
Cleff T (2014) Exploratory data analysis in business and economics. Explor Data Anal Bus Econ. https://doi.org/10.1007/978-3-319-01517-0
Dash S, Shakyawar SK, Sharma M, Kaushik S (2019) Big data in healthcare: management, analysis and future prospects. J Big Data 6(1):1–25
Davenport T, Kalakota R (2019) The potential for artificial intelligence in healthcare. Future Healthc J 6(2):94
Dsouza J (2020) Using exploratory data analysis for generating inferences on the correlation of COVID-19 cases. In: 2020 11th International Conference on Computing, Communication and Networking Technologies (ICCCNT). IEEE, pp 1–6
Faraggi D, Reiser B (2002) Estimation of the area under the ROC curve. Stat Med 21(20):3093–3106
Forte GF, Bauza JMT, de Pau V, Vall M, Camps A (2013) Experimental study on the performance of RFI detection algorithms in microwave radiometry: toward an optimum combined test. IEEE Trans Geosci Remote Sens 51(10):4936–4944
Garg M et al (2021) Computed tomography chest in COVID-19: when & why? Indian J Med Res 153(1–2):86
Habehh H, Gohel S (2021) Machine learning in healthcare. Curr Genomics 22(4):291–300
Hong SR, Hullman J, Bertini E (2020) Human factors in model interpretability: Industry practices, challenges, and needs. Proc ACM on Human-Comput Interact 4(CSCW1):1–26
Hulsen T et al (2019) From big data to precision medicine. Front Med 6:34
Hymer C, Smith AD (2022) Harnessing the positive side of negative cases: Exemplars and queries for qualitative researchers. Academy of management proceedings, 2022(1) Academy of Management Briarcliff Manor, NY 10510 Academy of Management, 202(1):14341
Jamwal S, Gautam A, Elsworth J, Kumar M, Chawla R, Kumar P (2020) An updated insight into the molecular pathogenesis, secondary complications and potential therapeutics of COVID-19 pandemic. Life Sci 257:118105
Karpievitch YV, Dabney AR, Smith RD (2012) Normalization and missing value imputation for label-free LC-MS analysis. BMC Bioinformatics 13(16):1–9
Kim ES et al (2020) Clinical course and outcomes of patients with severe acute respiratory syndrome coronavirus 2 infection: a preliminary report of the first 28 patients from the Korean cohort study on COVID-19. J Korean Med Sci 35(13):e142
La Rosa G, Bonadonna L, Lucentini L, Kenmoe S, Suffredini E (2020) Coronavirus in water environments: occurrence, persistence and concentration methods-A scoping review. Water Res 179:115899
Lei H-Y et al (2021) Potential effects of SARS-CoV-2 on the gastrointestinal tract and liver. Biomed Pharmacother 133:111064
Linnenbrink J, Milà C, Ludwig M, Meyer H (2023) kNNDM: k-fold nearest neighbour distance matching cross-validation for map accuracy estimation. Egusphere 2023:1–16
Magge A et al (2021) Proceedings of the sixth social media mining for health (#SMM4H) workshop and shared task. In: Proceedings of the sixth social media mining for health (# SMM4H) workshop and shared task
Maleki M, Mahmoudi MR, Wraith D, Pho K-H (2020) Time series modelling to forecast the confirmed and recovered cases of COVID-19. Travel Med Infect Dis 37:101742
Mehta N, Pandit A, Shukla S (2019) Transforming healthcare with big data analytics and artificial intelligence: a systematic mapping study. J Biomed Inform 100:103311
Mohamad IB, Usman D (2013) Standardization and its effects on K-means clustering algorithm. Res J Appl Sci Eng Technol 6(17):3299–3303
Munazhif NF, Yanris GJ, Hasibuan MNS (2023) Implementation of the K-nearest neighbor (kNN) method to determine outstanding student classes. Sinkron: Jurnal Dan Penelitian Teknik Informatika 8(2):719–732
Nadarajan R, Sulaiman N (2023) Evaluation of K-fold value in breast cancer diagnosis technique using SVM and bioinspired optimization algorithm (JA-ABC5). In: 2023 IEEE 13th symposium on computer applications & industrial electronics (ISCAIE). IEEE, pp 130–135
Nielsen SH et al (2021) 31,600-year-old human virus genomes support a Pleistocene origin for common childhood infections. BioRxiv. https://doi.org/10.1101/2021.06.28.450199
Oja E, Yuan Z (2006) The fastica algorithm revisited: convergence analysis. IEEE Trans Neural Netw 17(6):1370–1381
Ortiz-Prado E et al (2020) Clinical, molecular, and epidemiological characterization of the SARS-CoV-2 virus and the coronavirus disease 2019 (COVID-19), a comprehensive literature review. Diagn Microbiol Infect Dis 98(1):115094
Oyedele O (2023) Determining the optimal number of folds to use in a K-fold cross-validation: a neural network classification experiment. Res Math 10(1):2201015
Pandeva T, Forré P (2023) Multi-view independent component analysis with shared and individual sources. In: Uncertainty in artificial intelligence, PMLR, pp 1639–1650
Pattnayak P, Panda AR (2021) Innovation on machine learning in healthcare services—An introduction. IN: Technical advancements of machine learning in healthcare. Springer, pp 1–30
Pleil JD (2016) QQ-plots for assessing distributions of biomarker measurements and generating defensible summary statistics. J Breath Res 10(3):035001
Ramírez-Gallego S et al (2017) Fast-mRMR: fast minimum redundancy maximum relevance algorithm for high-dimensional big data. Int J Intell Syst 32(2):134–152
Ramosaj B, Pauly M (2023) Consistent and unbiased variable selection under indepedent features using random forest permutation importance. Bernoulli 29(3):2101–2118
Raoult D, Roux V (1997) Rickettsioses as paradigms of new or emerging infectious diseases. Clin Microbiol Rev 10(4):694–719
Sahlol AT, Yousri D, Ewees AA, Al-Qaness MA, Damasevicius R, Elaziz MA (2020) COVID-19 image classification using deep features and fractional-order marine predators algorithm. Sci Rep 10(1):1–15
Sakar CO, Kursun O, Gurgen F (2012) A feature selection method based on kernel canonical correlation analysis and the minimum redundancy-maximum relevance filter method. Expert Syst Appl 39(3):3432–3437
Schmidt JM, de Manuel M, Marques-Bonet T, Castellano S, Andrés AM (2019) Evidence that viruses, particularly SIV, drove genetic adaptation in natural populations of eastern chimpanzees. bioRxiv. https://doi.org/10.1101/582411
Sebe N, Lew MS, Cohen I, Garg A, Huang TS (2002) Emotion recognition using a cauchy naive bayes classifier. International conference on pattern recognition 1. IEEE, pp 17–20
Serrano CO et al (2020) Pediatric chest X-ray in covid-19 infection. Eur J Radiol 131:109236
Sethy PK, Behera SK (2020) Detection of coronavirus disease (covid-19) based on deep features
Sun X, Qourbani A (2023) Combining ensemble classification and integrated filter-evolutionary search for breast cancer diagnosis. J Cancer Res Clin Oncol 149(12):10753–10769
Tabaghi P, Khanzadeh M, Wang Y, Mirarab S (2023) Principal component analysis in space forms. arXiv preprint arXiv:2301.02750
Tebit DM et al (2020) Elucidating the viral and host factors enabling the cross-species transmission of primate lentiviruses from simians to humans. bioRxiv. https://doi.org/10.1101/2020.10.13.337303
Tsatsakis A et al (2020) SARS-CoV-2 pathophysiology and its clinical implications: an integrative overview of the pharmacotherapeutic management of COVID-19. Food Chem Toxicol 146:111769
Warren CJ, Sawyer SL (2023) Identifying animal viruses in humans. Science 379(6636):982–983
White J, Power SD (2023) k-fold cross-validation can significantly over-estimate true classification accuracy in common EEG-based passive BCI experimental designs: an empirical investigation. Sensors 23(13):6077
Woan Ching SL et al (2022) Multiclass convolution neural network for classification of COVID-19 CT images. Comput Intell Neurosci. https://doi.org/10.1155/2022/9167707
Xu Y et al (2021) Artificial intelligence: a powerful paradigm for scientific research. The Innovation 2(4):100179
Yachou Y, El Idrissi A, Belapasov V, Ait Benali S (2020) Neuroinvasion, neurotropic, and neuroinflammatory events of SARS-CoV-2: understanding the neurological manifestations in COVID-19 patients. Neurol Sci 41(10):2657–2669
Yang S, Rothman RE (2004) PCR-based diagnostics for infectious diseases: uses, limitations, and future applications in acute-care settings. Lancet Infect Dis 4(6):337–348
Zarzoso V, Comon P, Kallel M (2006) How fast is FastICA?. In: 2006 14th European signal processing conference. IEEE, pp 1–5
Acknowledgements
Finally, we express our gratitude to Dr. Mitra Esmaeili Azad, MD, from Shahid Beheshti University of Medical Sciences, for helping with the medical aspects of this research.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
Karlo Abnoosian and Rahman Farnoosh conceived the method. Karlo Abnoosian developed the algorithm and performed the simulations. Karlo Abnoosian and Rahman Farnoosh analysed the results and wrote the paper. All the authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests On behalf of all authors, the corresponding author states that there is no conflict of interest.
Ethics approval
This article is exempt and does not require ethics approval.
Consent to participate
Informed consent was obtained from all individual participants included in the study.
Consent to publish
The authors affirm that human research participants provided informed consent for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Farnoosh, R., Abnoosian, K. A robust innovative pipeline-based machine learning framework for predicting COVID-19 in Mexican patients. Int J Syst Assur Eng Manag (2024). https://doi.org/10.1007/s13198-024-02354-3
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13198-024-02354-3