Abstract
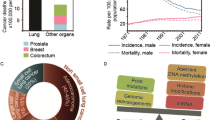
Lung cancer has a very high mortality in females and males. Most (~ 85%) of lung cancers are non-small cell lung cancers (NSCLC). When lung cancer is diagnosed, most of them have either local or distant metastasis, with a poor prognosis. In order to achieve better outcomes, it is imperative to identify the molecular signature based on genetic and epigenetic variations for different NSCLC subgroups. We hypothesize that DNA and histone modifications play significant roles in the framework of predictive, preventive, and personalized medicine (PPPM; 3P medicine). Epigenetics has a significant impact on tumorigenicity, tumor heterogeneity, and tumor resistance to chemotherapy, targeted therapy, and immunotherapy. An increasing interest is that epigenomic regulation is recognized as a potential treatment option for NSCLC. Most attention has been paid to the epigenetic alteration patterns of DNA and histones. This article aims to review the roles DNA and histone modifications play in tumorigenesis, early detection and diagnosis, and advancements and therapies of NSCLC, and also explore the connection between DNA and histone modifications and PPPM, which may provide an important contribution to improve the prognosis of NSCLC. We found that the success of targeting DNA and histone modifications is limited in the clinic, and how to combine the therapies to improve patient outcomes is necessary in further studies, especially for predictive diagnostics, targeted prevention, and personalization of medical services in the 3P medicine approach. It is concluded that DNA and histone modifications are potent diagnostic and therapeutic targets to advance non-small cell lung cancer management from the perspective of 3P medicine.



Similar content being viewed by others
Data availability
All data and materials are available in the current manuscript.
Abbreviations
- APC:
-
Adenomatous polyposis coli
- ALK:
-
Anaplastic lymphoma kinase
- CASP1:
-
Silenced caspase 1
- CNVs:
-
Copy number variations
- DSBs:
-
Double-strand breaks
- DNMTs:
-
DNA methyltransferases
- DNAm:
-
DNA methylation
- ELMO3:
-
Engulfment and cell motility 3
- EGFR:
-
Epidermal growth factor receptor
- FHIT:
-
Fragile histidine triad
- HR:
-
Homologous recombination
- hMLH1:
-
Human mutL homolog 1
- HRM:
-
Methylation-specific high-resolution melting
- HDACis:
-
Histone deacetylase inhibitors
- HATs:
-
Acetyltransferases
- KLF2:
-
Kruppel-like factor 2
- LUAD:
-
Lung adenocarcinoma
- LOH:
-
Loss of heterozygosity
- m6A:
-
N6-methyladenosine
- MMP:
-
Matrix metalloproteinase
- NSCLC:
-
Non-small cell lung cancer
- NHEJ:
-
Non-homologous end joining
- OG:
-
8-Oxo-7,8-dihydroguanine
- OS:
-
Overall survival
- PT:
-
Phosphorothioate
- PD-1:
-
Anti-programmed cell death protein 1
- PD-L1:
-
Anti-programmed cell death ligand 1
- PTMs:
-
Posttranslational modifications
- PFS:
-
Progression-free survival
- ROS:
-
Reactive oxygen species
- RARbeta:
-
Retinoic acid receptor-beta
- SMYD2:
-
SET and MYND domain-containing 2
- SCLC:
-
Small cell lung cancer
- TMB:
-
Tumor mutation burden
- TCGA:
-
The Cancer Genome Atlas
- TKIs:
-
Tyrosine kinase inhibitors
- TF:
-
Transcription factor
- TET:
-
Ten-eleven translocation enzymes
- TSGs:
-
Tumor suppressor genes
- 5mC:
-
5-Methylcytosine
- 5hmU:
-
5-Hydroxymethyluracil
- 5fU:
-
5-Formyluracil
- 5hmU:
-
5-Hydroxymethyluracil
- 5fU:
-
5-Formyluracil
- 5-Aza-dC:
-
5-Aza-2′-deoxycytidine
- 5-aza-CdR:
-
5-Aza-2′-deoxycytidine
References
Sung H, Ferlay J, Siegel RL. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA A Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. https://doi.org/10.21037/tlcr.2016.06.07.
Tamura T, Kurishima K, Nakazawa K, Kagohashi K, Ishikawa H, Satoh H, Hizawa N. Specific organ metastases and survival in metastatic non-small-cell lung cancer. Mole Clin Oncol. 2015;3(1):217–21. https://doi.org/10.3892/mco.2014.410.
Berger SL, Kouzarides T, Shiekhattar R, Shilatifard A. An operational definition of epigenetics. Genes Dev. 2009;23(7):781–3. https://doi.org/10.1101/gad.1787609.
Cheng T, Zhan X. Pattern recognition for predictive, preventive, and personalized medicine in cancer. EPMA J. 2017;8(1):51–60. https://doi.org/10.1007/s13167-017-0083-9.
Lu M, Zhan X. The crucial role of multiomic approach in cancer research and clinically relevant outcomes. EPMA J. 2018;9(1):77–102. https://doi.org/10.1007/s13167-018-0128-8.
Golubnitschaja O, Costigliola V. EPMA General report & recommendations in predictive, preventive and personalised medicine 2012: white paper of the European Association for Predictive Preventive Personalised Medicine. EPMA J. 2012;3(1):14. https://doi.org/10.1186/1878-5085-3-14.
Golubnitschaja O, Filep N, Yeghiazaryan K, Blom HJ, Hofmann-Apitius M, Kuhn W. Multi-omic approach decodes paradoxes of the triple-negative breast cancer: lessons for predictive, preventive and personalised medicine. Amino Acids. 2018;50(3–4):383–95. https://doi.org/10.1007/s00726-017-2524-0.
Greenberg MVC. Bourc’his D, The diverse roles of DNA methylation in mammalian development and disease. Nat Rev Mol Cell Biol. 2019;20(10):590–607. https://doi.org/10.1038/s41580-019-0159-6.
Koziol MJ, Bradshaw CR, Allen GE, Costa ASH, Frezza C, Gurdon JB. Identification of methylated deoxyadenosines in vertebrates reveals diversity in DNA modifications. Nat Struct Mol Biol. 2016;23(1):24–30. https://doi.org/10.1038/nsmb.3145.
Liu J, Zhu Y, Luo GZ, Wang X, Yue Y, Wang X, Zong X, Chen K, Yin H, Fu Y, et al. Abundant DNA 6mA methylation during early embryogenesis of zebrafish and pig. Nat Commun. 2016;7:13052. https://doi.org/10.1038/ncomms13052.
Wu TP, Wang T, Seetin MG, Lai Y, Zhu S, Lin K, Liu Y, Byrum SD, Mackintosh SG, Zhong M, et al. DNA methylation on N(6)-adenine in mammalian embryonic stem cells. Nature. 2016;532(7599):329–33. https://doi.org/10.1038/nature17640.
Hao Z, Wu T, Cui X, Zhu P, Tan C, Dou X, Hsu KW, Lin YT, Peng PH, Zhang LS, et al. N(6)-Deoxyadenosine methylation in mammalian mitochondrial DNA. Mol Cell. 2020;78(3):382-395.e388. https://doi.org/10.1016/j.molcel.2020.02.018.
Musheev MU. Baumgne, The origin of genomic N(6)-methyl-deoxyadenosine in mammalian cells. Nat Chem Biol. 2020;16(6):630–4. https://doi.org/10.1038/s41589-020-0504-2.
Liu X, Lai W, Li Y, Chen S, Liu B, Zhang N, Mo J, Lyu C, Zheng J, Du YR, et al. N(6)-methyladenine is incorporated into mammalian genome by DNA polymerase. Cell Res. 2021;31(1):94–7. https://doi.org/10.1038/s41422-020-0317-6.
Pfaffeneder T, Spada F, Wagner M, Brandmayr C, Laube SK, Eisen D, Truss M, Steinbacher J, Hackner B, Kotljarova O, et al. Tet oxidizes thymine to 5-hydroxymethyluracil in mouse embryonic stem cell DNA. Nat Chem Biol. 2014;10(7):574–81. https://doi.org/10.1038/nchembio.1532.
Roberts SA, Gordenin DA. Hypermutation in human cancer genomes: footprints and mechanisms. Nat Rev Cancer. 2014;14(12):786–800. https://doi.org/10.1038/nrc3816.
Yuan B, Jiang Y, Wang Y, Wang Y. Efficient formation of the tandem thymine glycol/8-oxo-7,8-dihydroguanine lesion in isolated DNA and the mutagenic and cytotoxic properties of the tandem lesions in Escherichia coli cells. Chem Res Toxicol. 2010;23(1):11–9. https://doi.org/10.1021/tx9004264.
Dai Y, Yuan BF, Feng YQ. Quantification and mapping of DNA modifications. RSC Chem Biol. 2021;2(4):1096–114. https://doi.org/10.1039/d1cb00022e.
Hotchkiss RD. The quantitative separation of purines, pyrimidines, and nucleosides by paper chromatography. J Biol Chem. 1948Aug;175(1):315–32.
Dor Y, Cedar H. Principles of DNA methylation and their implications for biology and medicine. Lancet. 2018;392(10149):777–86. https://doi.org/10.1016/s0140-6736(18)31268-6.
Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nat Rev Genet. 2002;3(6):415–28. https://doi.org/10.1038/nrg816.
Heyn H, Esteller M. An adenine code for DNA: a second life for N6-methyladenine. Cell. 2015;161(4):710–3. https://doi.org/10.1016/j.cell.2015.04.021.
Luo GZ, He C. DNA N(6)-methyladenine in metazoans: functional epigenetic mark or bystander? Nat Struct Mol Biol. 2017;24(6):503–6. https://doi.org/10.1038/nsmb.3412.
Chen T, Li E. Establishment and maintenance of DNA methylation patterns in mammals. Curr Top Microbiol Immunol. 2006;301:179–201. https://doi.org/10.1007/3-540-31390-7_6.
Kim H, Kwon YM, Kim JS, Han J, Shim YM, Park J, Kim DH. Elevated mRNA levels of DNA methyltransferase-1 as an independent prognostic factor in primary nonsmall cell lung cancer. Cancer. 2006;107(5):1042–9. https://doi.org/10.1002/cncr.22087.
Husni RE, Shiba-Ishii A, Iiyama S, Shiozawa T, Kim Y, Nakagawa T, Sato T, Kano J, Minami Y, Noguchi M. DNMT3a expression pattern and its prognostic value in lung adenocarcinoma. Lung Cancer. 2016;97:59–65. https://doi.org/10.1016/j.lungcan.2016.04.018.
Gao Q, Steine EJ, Barrasa MI, Hockemeyer D, Pawlak M, Fu D, Reddy S, Bell GW, Jaenisch R. Deletion of the de novo DNA methyltransferase Dnmt3a promotes lung tumor progression. Proc Natl Acad Sci U S A. 2011;108(44):18061–6. https://doi.org/10.1073/pnas.1114946108.
Tahiliani M, Koh KP, Shen Y, Pastor WA, Bandukwala H, Brudno Y, Agarwal S, Iyer LM, Liu DR, Aravind L, et al. Conversion of 5-methylcytosine to 5-hydroxymethylcytosine in mammalian DNA by MLL partner TET1. Science. 2009;324(5929):930–5. https://doi.org/10.1126/science.1170116.
Filipczak PT, Leng S, Tellez CS, Do KC, Grimes MJ, Thomas CL, Walton-Filipczak SR, Picchi MA, Belinsky SA. p53-suppressed oncogene TET1 prevents cellular aging in lung cancer. Cancer Res. 2019;79(8):1758–68. https://doi.org/10.1158/0008-5472.CAN-18-1234.
Rauch TA, Zhong X, Wu X, Wang M, Kernstine KH, Wang Z, Riggs AD, Pfeifer GP. High-resolution mapping of DNA hypermethylation and hypomethylation in lung cancer. Proc Natl Acad Sci U S A. 2008;105(1):252–7. https://doi.org/10.1073/pnas.0710735105.
Kalari S, Jung M, Kernstine KH, Takahashi T, Pfeifer GP. The DNA methylation landscape of small cell lung cancer suggests a differentiation defect of neuroendocrine cells. Oncogene. 2013;32(30):3559–68. https://doi.org/10.1038/onc.2012.362.
Brabender J, Usadel H, Danenberg KD, Metzger R, Schneider PM, Lord RV, Wickramasinghe K, Lum CE, Park J, Salonga D, et al. Adenomatous polyposis coli gene promoter hypermethylation in non-small cell lung cancer is associated with survival. Oncogene. 2001;20(27):3528–32. https://doi.org/10.1038/sj.onc.1204455.
Yu Q, Guo Q, Chen L, Liu S. Clinicopathological significance and potential drug targeting of CDH1 in lung cancer: a meta-analysis and literature review. Drug Des Dev Ther. 2015;9:2171–8. https://doi.org/10.2147/dddt.s78537.
Toyooka KO, Toyooka S, Virmani AK, Sathyanarayana UG, Euhus DM, Gilcrease M, Minna JD, Gazdar AF. Loss of expression and aberrant methylation of the CDH13 (H-cadherin) gene in breast and lung carcinomas. Can Res. 2001;61(11):4556–60.
Belinsky SA, Nikula KJ, Palmisano WA, Michels R, Saccomanno G, Gabrielson E, Baylin SB, Herman JG. Aberrant methylation of p16(INK4a) is an early event in lung cancer and a potential biomarker for early diagnosis. Proc Natl Acad Sci U S A. 1998;95(20):11891–6. https://doi.org/10.1073/pnas.95.20.11891.
Fong KM, Virmani AK, Geradts J, Gazdar AF, Minna JD. Aberrant promoter methylation of multiple genes in non-small cell lung cancers. Cancer Res. 2001;61(1):249–55.
Zhang Y, Xu R, Li G, Xie X, Long J, Wang H. Loss of expression of the differentially expressed in adenocarcinoma of the lung (DAL-1) protein is associated with metastasis of non-small cell lung carcinoma cells. Tumour Biol. 2012;33(6):1915–25. https://doi.org/10.1007/s13277-012-0452-x.
Yu J, Bulk E, Ji P, Hascher A, Tang M, Metzger R, Marra A, Serve H, Berdel WE, Wiewroth R, et al. The EPHB6 receptor tyrosine kinase is a metastasis suppressor that is frequently silenced by promoter DNA hypermethylation in non-small cell lung cancer. Clin Cancer Res. 2010;16(8):2275–83. https://doi.org/10.1158/1078-0432.Ccr-09-2000.
Hwang JA, Kim Y, Hong SH, Lee J, Cho YG, Han JY, Kim YH, Han J, Shim YM, Lee YS, et al. Epigenetic inactivation of heparan sulfate (glucosamine) 3-O-sulfotransferase 2 in lung cancer and its role in tumorigenesis. PLoS ONE. 2013;8(11):e79634. https://doi.org/10.1371/journal.pone.0079634.
Ma R, Feng N, Yu X, Lin H, Zhang X, Shi O, Zhang H, Zhang S, Li L, Zheng M, et al. Promoter methylation of Wnt/β-catenin signal inhibitor TMEM88 is associated with unfavorable prognosis of non-small cell lung cancer. Cancer Biol Med. 2017;14(4):377–86. https://doi.org/10.20892/j.issn.2095-3941.2017.0061.
Hashimoto K, Narita Y, Matsushita Y, Miyakita Y, Ono M, Kayama T, Shibui S. Methylation status of O6-methylguanine-DNA-methyl transferase promoter region in non-small-cell lung cancer patients with brain metastasis. Clin Transl Oncol. 2012;14(1):31–5. https://doi.org/10.1007/s12094-012-0758-6.
Wu F, Lu M, Qu L, Li DQ, Hu CH. DNA methylation of hMLH1 correlates with the clinical response to cisplatin after a surgical resection in Non-small cell lung cancer. Int J Clin Exp Pathol. 2015;8(5):5457–63.
Ibanez de Caceres I, Cortes-Sempere M, Moratilla C, Machado-Pinilla R, Rodriguez-Fanjul V, et al. IGFBP-3 hypermethylation-derived deficiency mediates cisplatin resistance in non-small-cell lung cancer. Oncogene. 2010;29(11):1681–90. https://doi.org/10.1038/onc.2009.454.
Fischer JR, Ohnmacht U, Rieger N, Zemaitis M, Stoffregen C, Manegold C, Lahm H. Prognostic significance of RASSF1A promoter methylation on survival of non-small cell lung cancer patients treated with gemcitabine. Lung Cancer. 2007;56(1):115–23. https://doi.org/10.1016/j.lungcan.2006.11.016.
Liu WB, Han F, Huang YS, Chen HQ, Chen JP, Wang DD, Jiang X, Yin L, Cao J, Liu JY. TMEM196 hypermethylation as a novel diagnostic and prognostic biomarker for lung cancer. Mol Carcinog. 2019;58(4):474–87. https://doi.org/10.1002/mc.22942.
Yao S, Wu D, Chen J, Wang P, Lv X, Huang J. Hypermethylation of the G protein-coupled receptor kinase 6 (GRK6) promoter inhibits binding of C/EBPα, and GRK6 knockdown promotes cell migration and invasion in lung adenocarcinoma cells. FEBS Open Bio. 2019;9(4):605–17. https://doi.org/10.1002/2211-5463.12606.
Toyooka S, Toyooka KO, Maruyama R, Virmani AK, Girard L, Miyajima K, Harada K, Ariyoshi Y, Takahashi T, Sugio K, et al. DNA methylation profiles of lung tumors. Mol Cancer Ther. 2001;1(1):61–7.
Z9.:61–67. S Lam, Geradts J, Ashfaq R, Virmani AK, Milchgrub S, Gazdar AF, Minna JD, 5′ CpG island methylation of the FHIT gene is correlated with loss of gene expression in lung and breast cancer. Cancer Res 2001, 61(9):3581–3585.
Gomes A, Reis-Silva M. Promoter hypermethylation of DNA repair genes MLH1 and MSH2 in adenocarcinomas and squamous cell carcinomas of the lung. Rev Port Pneumol. 2014;20(1):20–30. https://doi.org/10.1016/j.rppneu.2013.07.003.
Soria JC, Lee HY, Lee JI, Wang L, Issa JP, Kemp BL, Liu DD, Kurie JM, Mao L, Khuri FR. Lack of PTEN expression in non-small cell lung cancer could be related to promoter methylation. Clin Cancer Res. 2002;8(5):1178–84.
Sato K, Tomizawa Y, Iijima H, Saito R, Ishizuka T, Nakajima T, Mori M. Epigenetic inactivation of the RUNX3 gene in lung cancer. Oncol Rep. 2006;15(1):129–35.
Kuroki T, Trapasso F, Yendamuri S, Matsuyama A, Alder H, Williams NN, Kaiser LR, Croce CM. Allelic loss on chromosome 3p21.3 and promoter hypermethylation of semaphorin 3B in non-small cell lung cancer. Cancer Res. 2003;63(12):3352–5.
Virmani AK, Rathi A, Zsome 3p21.3 and promoter hypermethylation of semaphorin 3B in non-smallng KM, Thunnissen F et al. Promoter methylation and silencing of the retinoic acid receptor-beta gene in lung carcinomas. J Natl Cancer Inst 2000, 92(16):1303–1307. https://doi.org/10.1093/jnci/92.16.1303
Ponomaryova AA, Rykova EY, Cherdyntseva NV, Skvortsova TE, Dobrodeev AY. Zav’yalov AA, Tuzikov SA, Vlassov VV, Laktionov PP, RARβ2 gene methylation level in the circulating DNA from blood of patients with lung cancer. Eur J Cancer Prev. 2011;20(6):453–5. https://doi.org/10.1097/CEJ.0b013e3283498eb4.
Xu Z, Wang Y, Wang L, Xiong J, Wang H, Cui F, Peng H. The performance of the SHOX2/PTGER4 methylation assay is influenced by cancer stage, age, type and differentiation. Biomark Med. 2020;14(5):341–51. https://doi.org/10.2217/bmm-2019-0325.
Zhang HT, Chen XF, Wang MH, Wang JC, Qi QY, Zhang RM, Xu WQ, Fei QY, Wang F, Cheng QQ, et al. Defective expression of transforming growth factor beta receptor type II is associated with CpG methylated promoter in primary non-small cell lung cancer. Clin Cancer Res. 2004;10(7):2359–67. https://doi.org/10.1158/1078-0432.ccr-0959-3.
Fukami T, Fukuhara H, Kuramochi M, Maruyama T, Isogai K, Sakamoto M, Takamoto S, Murakami Y. Promoter methylation of the TSLC1 gene in advanced lung tumors and various cancer cell lines. Int J Cancer. 2003;107(1):53–9. https://doi.org/10.1002/ijc.11348.
Gainetdinov IV, Kapitskaya KY, Rykova EY, Ponomaryova AA, Cherdyntseva NV, Vlassov VV, Laktionov PP, Azhikina TL. Hypomethylation of human-specific family of LINE-1 retrotransposons in circulating DNA of lung cancer patients. Lung Cancer. 2016;99:127–30. https://doi.org/10.1016/j.lungcan.2016.07.005.
Søes S, Daugaard IL, Sørensen BS, Carus A, Mattheisen M, Alsner J, Overgaard J, Hager H, Hansen LL, Kristensen LS. Hypomethylation and increased expression of the putative oncogene ELMO3 are associated with lung cancer development and metastases formation. Oncoscience. 2014;1(5):367–74. https://doi.org/10.18632/oncoscience.42.
Yu J, Hou M, Pei T. FAM83A is a prognosis signature and potential oncogene of lung adenocarcinoma. DNA Cell Biol. 2020;39(5):890–9. https://doi.org/10.1089/dna.2019.4970.
Jang SJ, Soria JC, Wang L, Hassan KA, Morice RC, Walsh GL, Hong WK, Mao L. Activation of melanoma antigen tumor antigens occurs early in lung carcinogenesis. Cancer Res. 2001;61(21):7959–63.
Tsou JA, Hagen JA, Carpenter CL, Laird-Offringa IA. DNA methylation analysis: a powerful new tool for lung cancer diagnosis. Oncogene. 2002;21(35):5450–61. https://doi.org/10.1038/sj.onc.1205605.
Liang R, Li X, Li W, Zhu X, Li C. DNA methylation in lung cancer patients: Opening a “window of life” under precision medicine. Biomed Pharmacother. 2021;144:112202. https://doi.org/10.1016/j.biopha.2021.112202.
Ikeda K, Shiraishi K, Eguchi A, Shibata H, Yoshimoto K, Mori T, Baba Y, Baba H, Suzuki M. Long interspersed nucleotide element 1 hypomethylation is associated with poor prognosis of lung adenocarcinoma. Ann Thorac Surg. 2013;96(5):1790–4. https://doi.org/10.1016/j.athoracsur.2013.06.035.
Liu H, Liu W, Wu Y, Zhou Y, Xue R, Luo C, Wang L, Zhao W, Jiang JD, Liu J. Loss of epigenetic control of synuclein-gamma gene as a molecular indicator of metastasis in a wide range of human cancers. Cancer Res. 2005;65(17):7635–43. https://doi.org/10.1158/0008-5472.Can-05-1089.
Shao T, Song P, Hua H, Zhang H, Sun X, Kong Q, Wang J, Luo T, Jiang Y. Gamma synuclein is a novel Twist1 target that promotes TGF-β-induced cancer cell migration and invasion. Cell Death Dis. 2018;9(6):625. https://doi.org/10.1038/s41419-018-0657-z.
Smolle E, Pichler M, Non-smoking-associated lung cancer: a distinct entity in terms of tumor biology, patient characteristics and impact of hereditary cancer predisposition. Cancers (Basel) 2019, 11(2). https://doi.org/10.3390/cancers11020204
Gao X, Zhang Y, Breitling LP, Brenner H. Tobacco smoking and methylation of genes related to lung cancer development. Oncotarget. 2016;7(37):59017–28. https://doi.org/10.18632/oncotarget.10007.
Zhang Y, Elgizouli M. Smoking-associated DNA methylation markers predict lung cancer incidence. Clin Epigenetics. 2016;8:127. https://doi.org/10.1186/s13148-016-0292-4.
Baglietto L, Ponzi E, Haycock P, Hodge A, Bianca Assumma M, Jung CH, Chung J, Fasanelli F, Guida F, Campanella G, et al. DNA methylation changes measured in pre-diagnostic peripheral blood samples are associated with smoking and lung cancer risk. Int J Cancer. 2017;140(1):50–61. https://doi.org/10.1002/ijc.30431.
Toyooka S, Maruyama R, Toyooka KO, McLerran D, Feng Z, Fukuyama Y, Virmani AK, Zochbauer-Muller S, Tsukuda K, Sugio K, et al. Smoke exposure, histologic type and geography-related differences in the methylation profiles of non-small cell lung cancer. Int J Cancer. 2003;103(2):153–60. https://doi.org/10.1002/ijc.10787.
Kim DH, Nelson HH, Wiencke JK, Zheng S, Christiani DC, Wain JC, Mark EJ, Kelsey KT. p16(INK4a) and histology-specific methylation of CpG islands by exposure to tobacco smoke in non-small cell lung cancer. Cancer Res. 2001;61(8):3419–24.
Castro M, Grau L, Puerta P, Gimenez L, Venditti J, Quadrelli S. Multiplexed methylation profiles of tumor suppressor genes and clinical outcome in lung cancer. J Transl Med. 2010;8:86. https://doi.org/10.1186/1479-5876-8-86.
Damiani LA, Yingling CM, Leng S, Romo PE, Nakamura J, Belinsky SA. Carcinogen-induced gene promoter hypermethylation is mediated by DNMT1 and causal for transformation of immortalized bronchial epithelial cells. Cancer Res. 2008;68(21):9005–14. https://doi.org/10.1158/0008-5472.Can-08-1276.
Lin RK, Hsieh YS, Lin P, Hsu HS, Chen CY, Tang YA, Lee CF, Wang YC. The tobacco-specific carcinogen NNK induces DNA methyltransferase 1 accumulation and tumor suppressor gene hypermethylation in mice and lung cancer patients. J Clin Invest. 2010;120(2):521–32. https://doi.org/10.1172/jci40706.
O’Hagan HM, Wang W, Sen S, Destefano Shields C, Lee SS, Zhang YW, Clements EG, Cai Y, Van Neste L, Easwaran H, et al. Oxidative damage targets complexes containing DNA methyltransferases, SIRT1, and polycomb members to promoter CpG islands. Cancer Cell. 2011;20(5):606–19. https://doi.org/10.1016/j.ccr.2011.09.012.
Widschwendter M, Jones A, Evans I, Reisel D, Dillner J, et al. Epigenome-based cancer risk prediction: rationale, opportunities and challenges. Nat Rev Clin Oncol. 2018;15(5):292–309. https://doi.org/10.1038/nrclinonc.2018.30.
Ma Y, Bai Y, Mao H, Hong Q, Yang D, Zhang H, Liu F, Wu Z, Jin Q, Zhou H, et al. A panel of promoter methylation markers for invasive and noninvasive early detection of NSCLC using a quantum dots-based FRET approach. Biosens Bioelectron. 2016;85:641–8. https://doi.org/10.1016/j.bios.2016.05.067.
Ren M, Wang C, Sheng D, Shi Y, Jin M, Xu S. Methylation analysis of SHOX2 and RASSF1A in bronchoalveolar lavage fluid for early lung cancer diagnosis. Ann Diagn Pathol. 2017;27:57–61. https://doi.org/10.1016/j.anndiagpath.2017.01.007.
Ponomaryova AA, Rykova EY, Cherdyntseva NV, Skvortsova TE, Dobrodeev AY. Zav’yalov AA, Bryzgalov LO, Tuzikov SA, Vlassov VV, Laktionov PP, Potentialities of aberrantly methylated circulating DNA for diagnostics and post-treatment follow-up of lung cancer patients. Lung Cancer. 2013;81(3):397–403. https://doi.org/10.1016/j.lungcan.2013.05.016.
Kneip C, Schmidt B, Seegebarth A, Weickmann S, Fleischhacker M, Liebenberg V, Field JK, Dietrich D. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer in plasma. J Thorac Oncol. 2011;6(10):1632–8. https://doi.org/10.1097/JTO.0b013e318220ef9a.
Schmidt B, Liebenberg V, Dietrich D, Schlegel T, Kneip C, Seegebarth A, Flemming N, Seemann S, Distler J, Lewin J, et al. SHOX2 DNA methylation is a biomarker for the diagnosis of lung cancer based on bronchial aspirates. BMC Cancer. 2010;10:600. https://doi.org/10.1186/1471-2407-10-600.
Weiss G, Schlegel A, Kottwitz D. Validation of the SHOX2/PTGER4 DNA methylation marker panel for plasma-based discrimination between patients with malignant and nonmalignant lung disease. J Thorac Oncol. 2017;12(1):77–84. https://doi.org/10.1186/1471-2407-10-600.
Zhang C, Yu W, Wang L, Zhao M, Guo Q, Lv S, Hu X, Lou J. DNA methylation analysis of the SHOX2 and RASSF1A panel in bronchoalveolar lavage fluid for lung cancer diagnosis. J Cancer. 2017;8(17):3585–91. https://doi.org/10.7150/jca.21368.
Nunes SP, Diniz F, Moreira-Barbosa C, Constâncio V et al. Subtyping lung cancer using DNA methylation in liquid biopsies. J Clin Med 2019, 8 (9). https://doi.org/10.3390/jcm8091500
Grote HJ, Schmiemann V, Geddert H, Rohr UP, Kappes R, Gabbert HE. Aberrant promoter methylation of p16(INK4a), RARB2 and SEMA3B in bronchial aspirates from patients with suspected lung cancer. Int J Cancer. 2005;116(5):720–5. https://doi.org/10.1002/ijc.21090.
Zhao QT, Guo T, Wang HE, Zhang XP, Zhang H, Wang ZK, Yuan Z, Duan GC. Diagnostic value of SHOX2 DNA methylation in lung cancer: a meta-analysis. Onco Targets Ther. 2015;8:3433–9. https://doi.org/10.2147/ott.S94300.
Liu WJ, Tan XH, Guo BP, Ke Q, Sun J, Cen H. Associations between RASSF1A promoter methylation and NSCLC: a meta-analysis of published data. Asian Pac J Cancer Prev. 2013;14(6):3719–24. https://doi.org/10.7314/apjcp.2013.14.6.3719.
Walter RFH, Rozynek P, Casjens S, Werner R, Mairinger FD, Speel EJM, Zur Hausen A, Meier S, Wohlschlaeger J, Theegarten D, et al. Methylation of L1RE1, RARB, and RASSF1 function as possible biomarkers for the differential diagnosis of lung cancer. PLoS ONE. 2018;13(5):e0195716. https://doi.org/10.1371/journal.pone.0195716.
Ma Y, Li MD. Establishment of a strong link between smoking and cancer pathogenesis through DNA methylation analysis. Sci Rep. 2017;7(1):1811. https://doi.org/10.1038/s41598-017-01856-4.
Topper MJ, Vaz M, Chiappinelli KB, DeStefano Shields CE, Niknafs N, Yen RC, Wenzel A, Hicks J, Ballew M, Stone M, et al. Epigenetic therapy ties MYC depletion to reversing immune evasion and treating lung cancer. Cell. 2017;171(6):1284-1300.e1221. https://doi.org/10.1016/j.cell.2017.10.022.
Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377(25):2500–1. https://doi.org/10.1056/NEJMc1713444.
Cai L, Bai H, Duan J, Wang Z, Gao S, Wang D, Wang S, Jiang J, Han J, Tian Y, et al. Epigenetic alterations are associated with tumor mutation burden in non-small cell lung cancer. J Immunother Cancer. 2019;7(1):198. https://doi.org/10.1186/s40425-019-0660-7.
Wang W, Wang Q, Wan D, Sun Y, Wang L, Chen H, Liu C, Petersen RB, Li J, Xue W, et al. Histone HIST1H1C/H1.2 regulates autophagy in the development of diabetic retinopathy. Autophagy. 2017;13(5):941–54. https://doi.org/10.1080/15548627.2017.1293768.
Cheng X, Structural and functional coordination of DNA and histone methylation. Cold Spring Harb Perspect Biol 2014, 6(8). https://doi.org/10.1101/cshperspect.a018747
McGrath J, Trojer P. Targeting histone lysine methylation in cancer. Pharmacol Ther. 2015;150:1–22. https://doi.org/10.1016/j.pharmthera.2015.01.002.
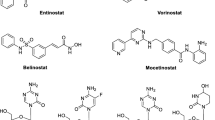
Damaskos C, Tomos I, Garmpis N, Karakatsani A, Dimitroulis D, Garmpi A, Spartalis E, Kampolis CF, Tsagkari E, Loukeri AA, et al. Histone deacetylase inhibitors as a novel targeted therapy against non-small cell lung cancer where are we now and what should we expect? Anticancer Res. 2018;38(1):37–43. https://doi.org/10.21873/anticanres.12189.
Audia JE, Campbell RM. Histone modifications and cancer. Cold Spring Harb Perspect Biol. 2016;8(4):a019521. https://doi.org/10.1101/cshperspect.a019521.
Petta V, Gkiozos I, Strimpakos A, Syrigos K. Histones and lung cancer: are the histone deacetylases a promising therapeutic target? Cancer Chemother Pharmacol. 2013;72(5):935–52. https://doi.org/10.1007/s00280-013-2223-9.
Li Z, Zhu WG. Targeting histone deacetylases for cancer therapy: from molecular mechanisms to clinical implications. Int J Biol Sci. 2014;10(7):757–70. https://doi.org/10.7150/ijbs.9067.
Glozak MA, Seto E. Histone deacetylases and cancer. Oncogene. 2007;26(37):5420–32. https://doi.org/10.1038/sj.onc.1210610.
Van Den Broeck A, Brambilla E, Moro-Sibilot D, Lantuejoul S, Brambilla C, Eymin B, Gazzeri S. Loss of histone H4K20 trimethylation occurs in preneoplasia and influences prognosis of non-small cell lung cancer. Clin Cancer Res. 2008;14(22):7237–45. https://doi.org/10.1158/1078-0432.Ccr-08-0869.
Bartling B, Hofmann HS, Boettger T, Hansen G, Burdach S, Silber RE, Simm A. Comparative application of antibody and gene array for expression profiling in human squamous cell lung carcinoma. Lung Cancer. 2005;49(2):145–54. https://doi.org/10.1016/j.lungcan.2005.02.006.
Osada H, Tatematsu Y, Saito H, Yatabe Y, Mitsudomi T, Takahashi T. Reduced expression of class II histone deacetylase genes is associated with poor prognosis in lung cancer patients. Int J Cancer. 2004;112(1):26–32. https://doi.org/10.1002/ijc.20395.
Minamiya Y, Ono T, Saito H, Takahashi N, Ito M, Motoyama S, Ogawa J. Strong expression of HDAC3 correlates with a poor prognosis in patients with adenocarcinoma of the lung. Tumour Biol. 2010;31(5):533–9. https://doi.org/10.1007/s13277-010-0066-0.
Riley JS, Hutchinson R, McArt DG, Crawford N, Holohan C, Paul I, Van Schaeybroeck S, Salto-Tellez M, Johnston PG, Fennell DA, et al. Prognostic and therapeutic relevance of FLIP and procaspase-8 overexpression in non-small cell lung cancer. Cell Death Dis. 2013;4(12):e951. https://doi.org/10.1038/cddis.2013.481.
Schrump DS, Fischette MR, Nguyen DM, Zhao M, Li X, Kunst TF, Hancox A, Hong JA, Chen GA, Kruchin E, et al. Clinical and molecular responses in lung cancer patients receiving Romidepsin. Clin Cancer Res. 2008;14(1):188–98. https://doi.org/10.1158/1078-0432.Ccr-07-0135.
Traynor AM, Dubey S, Eickhoff JC, Kolesar JM, Schell K, Huie MS, Groteluschen DL, Marcotte SM, Hallahan CM, Weeks HR, et al. Vorinostat (NSC# 701852) in patients with relapsed non-small cell lung cancer: a Wisconsin Oncology Network phase II study. J Thorac Oncol. 2009;4(4):522–6. https://doi.org/10.1097/jto.0b013e3181952478.
Reid T, Valone F, Lipera W, Irwin D, Paroly W, Natale R, Sreedharan S, Keer H, Lum B, Scappaticci F, et al. Phase II trial of the histone deacetylase inhibitor pivaloyloxymethyl butyrate (Pivanex, AN-9) in advanced non-small cell lung cancer. Lung Cancer. 2004;45(3):381–6. https://doi.org/10.1016/j.lungcan.2004.03.002.
Ryan QC, Headlee D, Acharya M, Sparreboom A, Trepel JB, Ye J, Figg WD, Hwang K, Chung EJ, Murgo A, et al. Phase I and pharmacokinetic study of MS-275, a histone deacetylase inhibitor, in patients with advanced and refractory solid tumors or lymphoma. J Clin Oncol. 2005;23(17):3912–22. https://doi.org/10.1200/jco.2005.02.188.
Prakash S, Foster BJ, Meyer M, Wozniak A, Heilbrun LK, Flaherty L, Zalupski M, Radulovic L, Valdivieso M, LoRusso PM. Chronic oral administration of CI-994: a phase 1 study. Invest New Drugs. 2001;19(1):1–11. https://doi.org/10.1023/a:1006489328324.
Ramalingam SS, Maitland ML, Frankel P, Argiris AE, Koczywas M, Gitlitz B, Thomas S, Espinoza-Delgado I, Vokes EE, Gandara DR, et al. Carboplatin and paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol. 2010;28(1):56–62. https://doi.org/10.1200/jco.2009.24.9094.
Tarhini AA, Zahoor H, McLaughlin B, Gooding WE, Schmitz JC, Siegfried JM, Socinski MA, Argiris A. Phase I trial of carboplatin and etoposide in combination with panobinostat in patients with lung cancer. Anticancer Res. 2013;33(10):4475–81.
Ramalingam SS, Parise RA, Ramanathan RK, Lagattuta TF, Musguire LA, Stoller RG, Potter DM, Argiris AE, Zwiebel JA, Egorin MJ, et al. Phase I and pharmacokinetic study of vorinostat, a histone deacetylase inhibitor, in combination with carboplatin and paclitaxel for advanced solid malignancies. Clin Cancer Res. 2007;13(12):3605–10. https://doi.org/10.1158/1078-0432.Ccr-07-0162.
Richards DA, Boehm KA, Waterhouse DM, Wagener DJ, Krishnamurthi SS, Rosemurgy A, Grove W, Macdonald K, Gulyas S, Clark M, et al. Gemcitabine plus CI-994 offers no advantage over gemcitabine alone in the treatment of patients with advanced pancreatic cancer: results of a phase II randomized, double-blind, placebo-controlled, multicenter study. Ann Oncol. 2006;17(7):1096–102. https://doi.org/10.1093/annonc/mdl081.
Pauer LR, Olivares J, Cunningham C, Williams A, Grove W, Kraker A, Olson S, Nemunaitis J. Phase I study of oral CI-994 in combination with carboplatin and paclitaxel in the treatment of patients with advanced solid tumors. Cancer Invest. 2004;22(6):886–96. https://doi.org/10.1081/cnv-200039852.
Chu BF, Karpenko MJ, Liu Z, Aimiuwu J, Villalona-Calero MA, Chan KK, Grever MR, Otterson GA. Phase I study of 5-aza-2′-deoxycytidine in combination with valproic acid in non-small-cell lung cancer. Cancer Chemother Pharmacol. 2013;71(1):115–21. https://doi.org/10.1007/s00280-012-1986-8.
Juergens RA, Wrangle J, Vendetti FP, Murphy SC, Zhao M, Coleman B, Sebree R, Rodgers K, Hooker CM, Franco N, et al. Combination epigenetic therapy has efficacy in patients with refractory advanced non-small cell lung cancer. Cancer Discov. 2011;1(7):598–607. https://doi.org/10.1158/2159-8290.Cd-11-0214.
Lin J, Gilbert J, Rudek MA, Zwiebel JA, Gore S, Jiemjit A, Zhao M, Baker SD, Ambinder RF, Herman JG, et al. A phase I dose-finding study of 5-azacytidine in combination with sodium phenylbutyrate in patients with refractory solid tumors. Clin Cancer Res. 2009;15(19):6241–9. https://doi.org/10.1158/1078-0432.Ccr-09-0567.
Candelaria M. Gallardo-Rincón A phase II study of epigenetic therapy with hydralazine and magnesium valproate to overcome chemotherapy resistance in refractory solid tumors. Ann Oncol. 2007;18(9):1529–38. https://doi.org/10.1093/annonc/mdm204.
Takeuchi S, Hase T, Shimizu S, Ando M, Hata A, Murakami H, Kawakami T, Nagase K, Yoshimura K, Fujiwara T, et al. Phase I study of vorinostat with gefitinib in BIM deletion polymorphism/epidermal growth factor receptor mutation double-positive lung cancer. Cancer Sci. 2020;111(2):561–70. https://doi.org/10.1111/cas.14260.
Witta SE, Jotte RM, Konduri K, Neubauer MA, Spira AI, Ruxer RL, Varella-Garcia M, Bunn PA Jr, Hirsch FR. Randomized phase II trial of erlotinib with and without entinostat in patients with advanced non-small-cell lung cancer who progressed on prior chemotherapy. J Clin Oncol. 2012;30(18):2248–55. https://doi.org/10.1200/jco.2011.38.9411.
Dasari A, Gore L, Messersmith WA, Diab S, Jimeno A, Weekes CD, Lewis KD, Drabkin HA, Flaig TW, Camidge DR. A phase I study of sorafenib and vorinostat in patients with advanced solid tumors with expanded cohorts in renal cell carcinoma and non-small cell lung cancer. Invest New Drugs. 2013;31(1):115–25. https://doi.org/10.1007/s10637-012-9812-z.
Reguart N, Rosell R, Cardenal F, Cardona AF, Isla D, Palmero R, Moran T, Rolfo C, Pallarexpanded coho, et al. Phase I/II trial of vorinostat (SAHA) and erlotinib for non-small cell lung cancer (NSCLC) patients with epidermal growth factor receptor (EGFR) mutations after erlotinib progression. Lung Cancer. 2014;84(2):161–7. https://doi.org/10.1016/j.lungcan.2014.02.011.
Gray JE, Haura E, Chiappori A, Tanvetyanon T, Williams CC, Pinder-Schenck M, Kish JA, Kreahling J, Lush R, Neuger A, et al. A phase I, pharmacokinetic, and pharmacodynamic study of panobinostat, an HDAC inhibitor, combined with erlotinib in patients with advanced aerodigestive tract tumors. Clin Cancer Res. 2014;20(6):1644–55. https://doi.org/10.1158/1078-0432.Ccr-13-2235.
Choi CYH, Wakelee HA, Neal JW, Pinder-Schenck MC, Yu HM, Chang SD, Adler JR, Modlin LA, Harsh GR, Soltys SG. Vorinostat and concurrent stereotactic radiosurgery for non-small cell lung cancer brain metastases: a phase 1 dose escalation trial. Int J Radiat Oncol Biol Phys. 2017;99(1):16–21. https://doi.org/10.1016/j.ijrobp.2017.04.041.
Gray JE, Saltos A, Tanvetyanon T, Haura EB, Creelan B, Antonia SJ, Shafique M, Zheng H, Dai W, Saller JJ, et al. Phase I/Ib study of pembrolizumab plus vorinostat in advanced/metastatic non-small cell lung cancer. Clin Cancer Res. 2019;25(22):6623–32. https://doi.org/10.1158/1078-0432.Ccr-19-1305.
Zuco V, De Cesare M, Cincinelli R, Nannei R, Pisano C, Zaffaroni N, Zunino F. Synergistic antitumor effects of novel HDAC inhibitors and paclitaxel in vitro and in vivo. PLoS ONE. 2011;6(12):e29085. https://doi.org/10.1371/journal.pone.0029085.
Groh T, Hrabeta J, Khalil MA, Doktorova H, Eckschlager T, Stiborova M. The synergistic effects of DNA-damaging drugs cisplatin and etoposide with a histone deacetylase inhibitor valproate in high-risk neuroblastoma cells. Int J Oncol. 2015;47(1):343–52. https://doi.org/10.3892/ijo.2015.2996.
Wang L, Li H, Ren Y, Zou S, Fang W, Jiang X, Jia L, Li M, Liu X, Yuan X, et al. Targeting HDAC with a novel inhibitor effectively reverses paclitaxel resistance in non-small cell lung cancer via multiple mechanisms. Cell Death Dis. 2016;7(1):e2063. https://doi.org/10.1038/cddis.2015.328.
Beg AA, Gray JE. HDAC inhibitors with PD-1 blockade: a promising strategy for treatment of multiple cancer types? Epigenomics. 2016;8(8):1015–7. https://doi.org/10.2217/epi-2016-0066.
Banik D, Moufarrij S, Villagra A, Immunoepigenetics combination therapies: an overview of the role of HDACs in cancer immunotherapy. Int J Mol Sci 2019, 20(9). https://doi.org/10.3390/ijms20092241
Wrangle J, Wang W, Koch A, Easwaran H, Mohammad HP, Vendetti F, Vancriekinge W, Demeyer T, Du Z, Parsana P, et al. Alterations of immune response of non-small cell lung cancer with azacytidine. Oncotarget. 2013;4(11):2067–79. https://doi.org/10.18632/oncotarget.1542.
Mehndiratta S, Lin MH, Wu YW, Chen CH, Wu TY, Chuang KH, Chao MW, Chen YY, Pan SL, Chen MC, et al. N-alkyl-hydroxybenzoyl anilide hydroxamates as dual inhibitors of HDAC and HSP90, downregulating IFN-γ induced PD-L1 expression. Eur J Med Chem. 2020;185:111725. https://doi.org/10.1016/j.ejmech.2019.111725.
Samuni Y, Wink DA, Krishna MC, Mitchell JB, Goldstein S. Suberoylanilide hydroxamic acid radiosensitizes tumor hypoxic cells in vitro through the oxidation of nitroxyl to nitric oxide. Free Radic Biol Med. 2014;73:291–8. https://doi.org/10.1016/j.freeradbiomed.2014.05.019.
Miller KM, Tjeertes JV, Coates J, Legube G, Polo SE, Britton S, Jackson SP. Human HDAC1 and HDAC2 function in the DNA-damage response to promote DNA nonhomologous end-joining. Nat Struct Mol Biol. 2010;17(9):1144–51. https://doi.org/10.1038/nsmb.1899.
Moore KE, Gozani O. An unexpected journey, lysine methylation across the proteome. Biochim Biophys Acta. 2014;1839(12):1395–403. https://doi.org/10.1016/j.bbagrm.2014.02.008.
Carlson SM, Moore KE, Green EM. Martation across the Proteome-wide enrichment of proteins modified by lysine methylation. Nat Protoc. 2014;9(1):37–50. https://doi.org/10.1038/nprot.2013.164.
Nguyen AT, Zhang Y. The diverse functions of Dot1 and H3K79 methylation. Genes Dev. 2011;25(13):1345–58. https://doi.org/10.1101/gad.2057811.
Nguyen AT, Taranova O, He J, Zhang Y. DOT1L, the H3K79 methyltransferase, is required for MLL-AF9-mediated leukemogenesis. Blood. 2011;117(25):6912–22. https://doi.org/10.1182/blood-2011-02-334359.
Xu C, Hao K, Hu H, Sheng Z, Yan J, Wang Q, Yu L. Expression of the enhancer of zeste homolog 2 in biopsy specimen predicts chemoresistance and survival in advanced non-small cell lung cancer receiving first-line platinum-based chemotherapy. Lung Cancer. 2014;86(2):268–73. https://doi.org/10.1016/j.lungcan.2014.09.010.
Wan D, Liu C, Sun Y, Wang W, Huang K, Zheng L. MacroH2A1.1 cooperates with EZH2 to promote adipogenesis by regulating Wnt signaling. J Mol Cell Biol. 2017;9(4):325–37. https://doi.org/10.1093/jmcb/mjx027.
Denissov S, Hofemeister H, Marks H, Kranz A, Ciotta G, Singh S, Anastassiadis K, Stunnenberg HG, Stewart AF. Mll2 is required for H3K4 trimethylation on bivalent promoters in embryonic stem cells, whereas Mll1 is redundant. Development. 2014;141(3):526–37. https://doi.org/10.1242/dev.102681.
Yin S, Yang J, Lin B, Deng W, Zhang Y, Yi X, Shi Y, Tao Y, Cai J, Wu CI, et al. Exome sequencing identifies frequent mutation of MLL2 in non-small cell lung carcinoma from Chinese patients. Sci Rep. 2014;4:6036. https://doi.org/10.1038/srep06036.
Xue W, Huang J, Chen H, Zhang Y, Zhu X, Li J, Zhang W, Yuan Y, Wang Y, Zheng L, et al. Histone methyltransferase G9a modulates hepatic insulin signaling via regulating HMGA1. Biochim Biophys Acta Mol Basis Dis. 2018;1864(2):338–46. https://doi.org/10.1016/j.bbadis.2017.10.037.
Huang T, Zhang P, Li W, Zhao T, Zhang Z, Chen S, Yang Y, Feng Y, Li F, Shirley Liu X, et al. G9A promotes tumor cell growth and invasion by silencing CASP1 in non-small-cell lung cancer cells. Cell Death Dis. 2017;8(4):e2726. https://doi.org/10.1038/cddis.2017.65.
Wang R, Deng X, Yoshioka Y, Vougiouklakis T, Park JH, Suzuki T, Dohmae N, Ueda K, Hamamoto R, Nakamura Y. Effects of SMYD2-mediated EML4-ALK methylation on the signaling pathway and growth in non-small-cell lung cancer cells. Cancer Sci. 2017;108(6):1203–9. https://doi.org/10.1111/cas.13245.
Hao C, Wang L, Peng S, Cao M, Li H, Hu J, Huang X, Liu W, Zhang H, Wu S, et al. Gene mutations in primary tumors and corresponding patient-derived xenografts derived from non-small cell lung cancer. Cancer Lett. 2015;357(1):179–85. https://doi.org/10.1016/j.canlet.2014.11.024.
Walter DM, Venancio OS, Buza EL, Tobias JW, Deshpande C, Gudiel AA, Kim-Kiselak C, Cicchini M, Yates TJ, Feldser DM. Systematic in vivo inactivation of chromatin-regulating enzymes identifies Setd2 as a potent tumor suppressor in lung adenocarcinoma. Cancer Res. 2017;77(7):1719–29. https://doi.org/10.1158/0008-5472.Can-16-2159.
Takawa M, Cho HS, Hayami S, Toyokawa G, Kogure M, Yamane Y, Iwai Y, Maejima K, Ueda K, Masuda A, et al. Histone lysine methyltransferase SETD8 promotes carcinogenesis by deregulating PCNA expression. Cancer Res. 2012;72(13):3217–27. https://doi.org/10.1158/0008-5472.Can-11-3701.
Chen T, Ren H, Thakur A, Yang T, Li Y, Zhang S, Wang T, Chen M. miR-382 inhibits tumor progression by targeting SETD8 in non-small cell lung cancer. Biomed Pharmacother. 2017;86:248–53. https://doi.org/10.1016/j.biopha.2016.12.007.
Lv T, Yuan D, Miao X, Lv Y, Zhan P, Shen X, Song Y. Over-expression of LSD1 promotes proliferation, migration and invasion in non-small cell lung cancer. PLoS ONE. 2012;7(4):e35065. https://doi.org/10.1371/journal.pone.0035065.
He R, Zhang FH, Shen N. LncRNA FEZF1-AS1 enhances epithelial-mesenchymal transition (EMT) through suppressing E-cadherin and regulating WNT pathway in non-small cell lung cancer (NSCLC). Biomed Pharmacother. 2017;95:331–8. https://doi.org/10.1016/j.biopha.2017.08.057.
Kong L, Zhang P, Li W, Yang Y, Tian Y, Wang X, Chen S, Yang Y, Huang T, Zhao T, et al. KDM1A promotes tumor cell invasion by silencing TIMP3 in non-small cell lung cancer cells. Oncotarget. 2016;7(19):27959–74. https://doi.org/10.18632/oncotarget.8563.
Mosammaparast N, Shi Y. Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu Rev Biochem. 2010;79:155–79. https://doi.org/10.1146/annurev.biochem.78.070907.103946.
Wagner KW, Alam H, Dhar SS, Giri U, Li N, Wei Y, Giri D, Cascone T, Kim JH, Ye Y, et al. KDM2A promotes lung tumorigenesis by epigenetically enhancing ERK1/2 signaling. J Clin Invest. 2013;123(12):5231–46. https://doi.org/10.1172/jci68642.
Dhar SS, Alam H, Li N, Wagner KW, Chung J, Ahn YW, Lee MG. Transcriptional repression of histone deacetylase 3 by the histone demethylase KDM2A is coupled to tumorigenicity of lung cancer cells. J Biol Chem. 2014;289(11):7483–96. https://doi.org/10.1074/jbc.M113.521625.
Cho HS, Toyokawa G, Daigo Y, Hayami S, Masuda K, Ikawa N, Yamane Y, Maejima K, Tsunoda T, Field HI, et al. The JmjC domain-containing histone demethylase KDM3A is a positive regulator of the G1/S transition in cancer cells via transcriptional regulation of the HOXA1 gene. Int J Cancer. 2012;131(3):E179-189. https://doi.org/10.1002/ijc.26501.
Zhan M, Wen F, Liu L, Chen Z, Wei H, Zhou H. JMJD1A promotes tumorigenesis and forms a feedback loop with EZH2/let-7c in NSCLC cells. Tumour Biol. 2016;37(8):11237–47. https://doi.org/10.1007/s13277-016-4999-9.
Wu Q, Tian Y, Zhang J, Tong X, Huang H, Li S, Zhao H, Tang Y, Yuan C, Wang K, et al. In vivo CRISPR screening unveils histone demethylase UTX as an important epigenetic regulator in lung tumorigenesis. Proc Natl Acad Sci U S A. 2018;115(17):E3978-e3986. https://doi.org/10.1073/pnas.1716589115.
Iderzorig T, Kellen J, Osude C, Singh S, Woodman JA, Garcia C, Puri N. Comparison of EMT mediated tyrosine kinase inhibitor resistance in NSCLC. Biochem Biophys Res Commun. 2018;496(2):770–7. https://doi.org/10.1016/j.bbrc.2018.01.069.
Zhang J, Ni SS, Zhao WL, Dong XC, Wang JL. High expression of JMJD6 predicts unfavorable survival in lung adenocarcinoma. Tumour Biol. 2013;34(4):2397–401. https://doi.org/10.1007/s13277-013-0789-9.
Duan Q, Pang C, Chang N, Zhang J, Liu W. Overexpression of PAD4 suppresses drug resistance of NSCLC cell lines to gefitinib through inhibiting Elk1-mediated epithelial-mesenchymal transition. Oncol Rep. 2016;36(1):551–8. https://doi.org/10.3892/or.2016.4780.
Kikuchi J, Takashina T, Kinoshita I, Kikuchi E, Shimizu Y, Sakakibara-Konishi J, Oizumi S, Marquez VE, Nishimura M, Dosaka-Akita H. Epigenetic therapy with 3-deazaneplanocin A, an inhibitor of the histone methyltransferase EZH2, inhibits growth of non-small cell lung cancer cells. Lung Cancer. 2012;78(2):138–43. https://doi.org/10.1016/j.lungcan.2012.08.003.
Fillmore CM, Xu C, Desai PT, Berry JM, Rowbotham SP, Lin YJ, Zhang H, Marquez VE, Hammerman PS, Wong KK, et al. EZH2 inhibition sensitizes BRG1 and EGFR mutant lung tumours to TopoII inhibitors. Nature. 2015;520(7546):239–42. https://doi.org/10.1038/nature14122.
Kim W, Kim R, Park G, Park JW, Kim JE. Deficiency of H3K79 histone methyltransferase Dot1-like protein (DOT1L) inhibits cell proliferation. J Biol Chem. 2012;287(8):5588–99. https://doi.org/10.1074/jbc.M111.328138.
Tan J, Yang X, Zhuang L, Jiang X, Chen W, Lee PL, Karuturi RK, Tan PB, Liu ET, Yu Q. Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev. 2007;21(9):1050–63. https://doi.org/10.1101/gad.1524107.
McCabe MT, Ott HM, Ganji G, Korenchuk S, Thompson C, Van Aller GS, Liu Y, Graves AP, Della Pietra A, Diaz E, et al. EZH2 inhibition as a therapeutic strategy for lymphoma with EZH2-activating mutations. Nature. 2012;492(7427):108–12. https://doi.org/10.1038/nature11606.
Gulati N, Béguelin W, Giulino-Roth L. Enhancer of zeste homolog 2 (EZH2) inhibitors. Leuk Lymphoma. 2018;59(7):1574–85. https://doi.org/10.1080/10428194.2018.1430795.
Vaswani RG, Gehling VS, Dakin LA, Cook AS, Nasveschuk CG, Duplessis M, Iyer P, Balasubramanian S, Zhao F, Good AC, et al. Identification of (R)-N-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-2-methyl-1-(1-(1-(2,2,2-trifluoroethyl)piperidin-4-yl)ethyl)-1H-indole-3-carboxamide (CPI-1205), a potent and selective inhibitor of histone methyltransferase EZH2, suitable for phase I clinical trials for B-cell lymphomas. J Med Chem. 2016;59(21):9928–41. https://doi.org/10.1021/acs.jmedchem.6b01315.
Stewart CA, Byers LA. Altering the course of small cell lung cancer: targeting cancer stem cells via LSD1 inhibition. Cancer Cell. 2015;28(1):4–6. https://doi.org/10.1016/j.ccell.2015.06.011.
Zhan X, Li J, Guo Y, Golubnitschaja O. Mass spectrometry analysis of human tear fluid biomarkers specific for ocular and systemic diseases in the context of 3P medicine. EPMA J. 2021;12(4):449–75. https://doi.org/10.1007/s13167-021-00265-y.
Li N, Desiderio DM, Zhan X. The use of mass spectrometry in a proteome-centered multiomics study of human pituitary adenomas. Mass Spectrom Rev. 2022;41(6):964–1013. https://doi.org/10.1002/mas.21710.
Acknowledgements
The authors acknowledge the financial support from the Shandong First Medical University Talent Introduction Funds (to X.Z.), Shandong First Medical University High-level Scientific Research Achievement Cultivation Funding Program (to X.Z.), the Shandong Provincial Natural Science Foundation (ZR202103020356/ZR2021MH156 to X.Z.), Taishan Scholar Engineering Project Special Funds (to X.Z.), and the Academic Promotion Program of Shandong First Medical University (2019ZL002).
Funding
This work was supported by the Shandong First Medical University Talent Introduction Funds (to X.Z.), Shandong First Medical University High-level Scientific Research Achievement Cultivation Funding Program (to X.Z.), the Shandong Provincial Natural Science Foundation (ZR202103020356/ZR2021MH156 to X.Z.), Taishan Scholar Engineering Project Special Funds (to X.Z.), and the Academic Promotion Program of Shandong First Medical University (2019ZL002).
Author information
Authors and Affiliations
Contributions
G.Z. and Z.W. collected and analyzed literature, wrote the manuscript, they contributed equally to the manuscript. P.S. participated in collection and analysis of literature. X.Z. conceived the concept, designed the manuscript, coordinated, and critically revised the manuscript, and was responsible for the corresponding works. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, G., Wang, Z., Song, P. et al. DNA and histone modifications as potent diagnostic and therapeutic targets to advance non-small cell lung cancer management from the perspective of 3P medicine. EPMA Journal 13, 649–669 (2022). https://doi.org/10.1007/s13167-022-00300-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13167-022-00300-6