Abstract
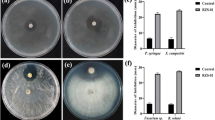
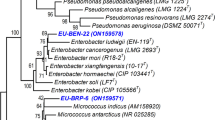
The consistent application of agrochemical herbicides has been reported to impact negatively on human health, environment, and food safety, and facilitated the emergence of weed resistances. Rhizosphere bacteria (RB) of different crops were screened for antagonism against Amaranthus hybridus L. (pigweed) and Echinochloa crus-galli (L.) Beauv. (barnyard grass) using necrosis assay technique. A total of eight rhizosphere bacterial isolates (B1–B8) produced different degrees of leaf necrosis on target weeds with isolate B2 manifesting the most significant necrotic activity. The rhizospheric bacterium (B2) with the highest necrotic activity was identified using 16S rRNA sequencing technique and further investigated. Molecular, morphological, and biochemical characterizations confirmed B2 isolate to be Pseudomonas aeruginosa. On isolation with ethyl acetate, separation, defatting, purification, and flash chromatography, seven different fractions (fraction 1–fraction 7) were obtained out of which fraction 4 showed the highest necrotic activity in necrosis assay experiment. Preparative HPLC of fraction 4 resulted in a pure compound that completely inhibited seed germination and seedling development of pigweed and barnyard grass but remained non-antagonistic to other tested soil fungi used in this study. The result obtained from this present study consequently confirmed the antagonistic behavior of rhizosphere-inhabiting P. aeruginosa to the target weeds and qualified the suitability of bacterium as good alternative source of bioherbicide. Potential herbicidal formulation from P. aeruginosa will help reduce crop loss due to weed challenges while offering a partial solution to the use of agrochemicals and food security.

ᅟ





Similar content being viewed by others
References
Abouziena HFH, Omar AAM, Sharma SD, Singh M (2009) Efficacy comparison of some new natural product herbicides for weed control at two growth stages. Weed Technol 23(3):431–437
Adeosun JO, Dauda CK, Gezui MA, Odunze AC, Amapu IY, Kudp T (2009) On-farm weed management in upland rice in three villages of Katsina State of Nigeria. African Crop Science Society. Afr Crop Sci Conf Proc 9:625–629
Adetunji CO, Oloke JK (2013) Efficacy of freshly prepared pesta granular formulations from the multi-combination of wild and mutant strain of Lasiodiplodia pseudotheobromae and Pseudomonas aeruginosa. Albanian J Agric Sci 12(4):555–563
Adigun JA, Lagoke STO (2003) Assessment of critical period of weed interference in transplanted rainfed and irrigated tomatoes in the Nigerian Northern Guinea Savanna. Niger J Plant Prot 21:89–100
Adigun J, Osipitan AO, Lagoke ST, Adeyemi RO, Afolami SO (2014) Growth and yield performance of cowpea (Vigna unguiculata (L.) Walp) as influenced by row-spacing and period of weed interference in South-West Nigeria. J Agric Sci 6(4):188–198
Al-Hinai AH, Al-Sadi AM, Al-Bahry MAS, Al-said FA, Al-Harthi SA, Deadman ML (2010) Isolation and characterization of Pseudomonas aeruginosa with antagonistic activity against Pythium aphanidermatum. J Plant Pathol 92(3):653–660
Allen VB, Randall GP (2014) Management of vegetation by alternative practices in fields and roadsides. Int J Agron 2014:1–12. doi:10.1155/2014/207828
Altschul SF (1997) Gapped BLAST & PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25(17):3389–3402
Asiwe J, Kutu RF (2007) Effect of plant spacing on yield weeds insect infestation and leaf bright of Bambara groundnut. Proc Afr Crop Sci Soc 4:1947–1950
Baghestani MA, Zand E, Soufizadeh S, Bagherani N, Deihimfard R (2007) Weed control and wheat (Triticum aestivum L.) yield under application of 2, 4-D plus carfentrazone-ethyl and florasulam plus flumetsulam: evaluation of the efficacy. Crop Prot 26(12):1759–1764
Bolton H, Elliott LF (1989) Toxin production by a rhizobacterial Pseudomonas sp. that inhibits wheat root growth. Plant Soil 114(2):269–278
Brainard DC, Curran WS, Bellinder RR (2013) Temperature and relative humidity affect weed response to vinegar and clove oil. Weed Technol 27(1):156–164
Caldwell CJ, Hynes RK, Boyetchko SM, Korber DR (2012) Colonization and bioherbicidal activity on green foxtail by Pseudomonas fluorescens BRG100 in a pesta formulation. Can J Microbiol 58(1):1–9
Cappuccino JG, Sherman N (1992) Microbiology, a laboratory manual. The Benjamin Cummings Publishing Company Inc., California, p 462
Cattelan AJ, Hartel PG, Fuhrmann JJ (1999) Screening for plant growth-promoting rhizobacteria to promote early soybean growth. Soil Sci Soc Am J 63(6):1670–1680
Charudattan R (1991) The mycoherbicide approach with plant pathogens. In: TeBeest DO (ed) Microbial control of weeds. Chapman & Hall, New York, pp 24–57. doi:10.1007/978-1-4615-9680-6_2
Chikoye D, Schulz S, Ekeleme F (2004) Evaluation of integrated weed management practices for maize in the northern guinea savanna of Nigeria. Crop Prot 23(10):895–900
Crump NS, Ash GJ, Fagan RJ (1999) The development of an Australian bioherbicide. 12 Australian Weed Conference, pp 235–237
Dueñas M, González-Manzano S, Surco-Laos F, González-Paramas A, Santos-Buelga C (2012) Characterization of sulfated quercetin and epicatechin metabolites. J Agric Food Chem 60(14):3592–3598
Ekeleme E, Kamara AY, Oikeh SO, Chikoye D, Omoigui LO (2007) Effect of weed competition on upland rice production in north-eastern Nigeria. Afr Crop Sci Conf Proc 8:61–65
Evidente A, Motta A (2001) Phytotoxins from fungi, pathogenic for agrarian forestal and weedy plants. In: Tringali C (ed) Bioactive compounds from natural source. Taylor& Francis, London, pp 473–525
FAOSTAT (2013) Food and agricultural organization of the United States. Date retrieved, 27th March, 2017. http://faostat.fao.org/site/567/default
Fawole MO, Oso BA (2004) Biochemical test. Laboratory manual of microbiology. Spectrum Books Limited, Ibadan, pp 14–17
Flores-Vargas RD, O’Hara GW (2006) Isolation and characterization of rhizosphere bacteria with potential for biological control of weeds in vineyards. J Appl Microbiol 100(5):946–954
Gurusiddaiah S, Gealy DR, Kennedy AC, Ogg AG (1994) Isolation and characterization of metabolites from Pseudomonas fluorescens-D7 for control of downy brome (Bromus tectorum). Weed Sci 42:492–501
Hamid AA, Aiyelaagbe OO, Balogun GA (2011) Herbicides and its applications. Adv Nat Appl Sci 5(2):201–213
Harding DP, Raizada MN (2015) Controlling weeds with fungi, bacteria and viruses: a review. Front Plant Sci 6:1–14
Heap I (2005) The international survey of herbicide resistant weeds. Web page: www.weedscience.com
Ivany JA (2010) Acetic acid for weed control in potato (Solanum tuberosum L.) Can J Plant Sci 90(4):537–542
Javaid A (2010) Herbicidal potential of allelopathic plants and fungi against Parthenium hysterophorus—a review. Allelopath J 25(2):331–344
Javaid A, Adrees H (2009) Parthenium management by cultural filtrates of phytopathogenic fungi. Nat Prod Res 23(16):1541–1551
Javaid A, Ali S (2011) Herbicidal activity of culture filtrates of Trichoderma spp. against two problematic weeds of wheat. Nat Prod Res 25(7):730–740
Javaid A, Shafique S, Shafique S (2010) Herbicidal effects of extracts and residue incorporation of Daturametelagainst partheniumweed. Nat Prod Res 24(15):1426–1437
Javaid A, Shafique S, Shafique S (2011) Management of Parthenium hysterophorus (Asteraceae) by Withania somnifera (Solanaceae). Nat Prod Res 25(4):407–416
Johnson WC III, Boudreau MA, Davis JW (2013) Combinations of corn gluten meal, clove oil, and sweep cultivation are ineffective for weed control in organic peanut production. Weed Technol 27(2):417–421
Kaewchai S, Soytong K, Hyde KD (2009) Mycofungicides and fungal biofertilizers. Fungal Divers 38:25–50. http://www.fungaldiversity.org/fdp/sfdp/FD38-2.pdf
Kennedy AC, Stubbs TL (2007) Management effects on the incidence of jointed goat grass inhibitory rhizobacteria. Biol Control 40(2):213–221
Kennedy AC, Young FL, Elliott LF, Douglas CL (1991) Rhizobacteria suppressive to the weed downy brome. Soil Sci Soc Am J 55(3):722–727
Kloepper JW, Schroth MN, Miller TD (1980) Effects of Rhizosphere Colonization by Plant Growth-Promoting Rhizobacteria on Potato Plant Development and Yield. Phytopathology 70(11):1078-1082
Kordali S, Cakir A, Sutay S (2007) Inhibitory effects of monoterpenes on seed germination and seedling growth. Zeitschrift fur Naturforschung (C) 62(3):207–214
Kordali S, Cakir A, Ozer H, Cakmakci R, Kesdek M, Mete E (2008) Antifungal, phytotoxic and insecticidal properties of essential oil isolated from Turkish Origanum acutidens and its three components, carvacrol, thymol and p-cymene. Bioresour Technol 99(18):8788–8795
Kremer RJ (2005) The role of bioherbicides in weed management. Biopestic Int 1(3,4):127–141
Kremer RJ, Kennedy AC (1996) Rhizobacteria as biocontrol agents of weeds. Weed Technol 10:601–609
Kremer RJ, Begonia MFT, Stanley L, Lanham ET (1990) Characterization of rhizobacteria associated with weed seedlings. Appl Environ Microbiol 56(4):1649–1655
Lugtenberg BJJ, Dekkers LC (1999) What make Pseudomonas bacteria rhizosphere competent? Environ Microbiol 1(1):9–13
Makhan SB, Simerjit K, Tarundeep K, Tarlok S, Megh S, Amit JJ (2013) Control of broadleaf weeds with post-emergence herbicides in four barley (Hordeum spp.) cultivars. Crop Prot 43:216–222. www.sciencedirect.com/science/article/pii/S0261219412002785
Mark R, Behrens NM, Sarbani C, Razvan D, Wen Z, Bradley J, LaVallee PL, Herman TE, Clemente DP (2007) Weeks dicamba resistance: enlarging and preserving biotechnology-based weed management strategies. Science 316(5828):1185–1188. doi:10.1126/science.1141596
McPhail KL, Armstrong DJ, Azevedo MD, Banowetz GM, Mills DI (2010) 4-Formylaminooxyvinylglycine, an herbicidal germination arrest factor (GAF) from Pseudomonas rhizosphere bacteria. J Nat Prod 73(11):1853–1857
Mejri D, Gamalero E, Souissi T (2013) Formulation development of the deleterious rhizobacterium Pseudomonas trivialisX33d for biocontrol of brome (Bromus diandrus) in durum wheat. J Appl Microbiol 114(1):219–228. doi:10.1111/jam.12036
Muhammad AS, Muhammad M, Muhammad AUL, Abid N (2012) Efficacy of various herbicides against weeds in wheat (Triticum aestivum L.) Afr J Biotechnol 11(4):791–799. doi:10.5897/AJB11.3274
Nurse RE, Hamill AS, Swanton CJ, Tardif FJ, Sikkema PH (2007) Weed control and yield response to foramsulfuron in corn. Weed Technol 21(2):453–458
Poston DH, Wilson HP, Hines TE (2000) Imidazolinone resistance in several Amaranthus hybridus populations. Weed Sci 48(4):508–513
Sacchi CT, Whitney AM, Mayer LW, Morey R, Steigerwalt A, Boras A, Weyant RS, Popovic T (2002) Sequencing of 16S rRNA gene: a rapid tool for identification of Bacillus anthracis. Emerg Infect Dis 8(10):1117–1123
Schippers B, Bakker AW, Bakker PAHM (1987) Interactions of deleterious and beneficial rhizosphere microorganisms and the effect of cropping practices. Annu Rev Phytopathol 25:339–358
Sessitsch A, Gyamfi S, Tscherko D, Gerzabek MH, Kandeler E (2004) Activity of microorganisms in the rhizosphere of herbicide treated and untreated transgenic glufosinate-tolerant and wild type oilseeds rape grown in containment. Plant Soil 266(1):105–116. doi:10.1007/s11104-005-7077-4
Takim FO, Amodu AA (2013) Quantitative estimate of weeds of sugarcane (Saccharum officinarum L.) crop in Ilorin, southern Guinea savannah of Nigeria. Ethiop J Environ Stud Manag 6(6):611–619
Takim FO, Ahmadu MS, Omotosho SB (2015) Efficacy of Ametryn herbicides on weeds of sugarcane in southern guinea savanna zone of Nigeria. Niger J Agric, Food Environ 11(3):147–151
Usman A, Elemo KA, Lagoke STO, Adigun JA (2002) Nitrogen and weed management in maize intercropped with upland rice. J Sustain Agric 21(1):5–16. doi:10.1300/J064v21n01_03
Van Driesche RG, Carruthers RI, Center T, Hoddle MS, Hough-Goldstein J, Morin L, Smith DL (2010) Classical biological control for the protection of natural ecosystem. Biol Control 154(S1):2–33
Xiaoya C, Mengmeng G (2016) Bioherbicides in organic agriculture. Horticulturae 2(3):1–10
Yang J, Hong-zhe CAO, Wang WV (2014) Isolation, identification, and herbicidal activity of metabolites produced by Pseudomonas aeruginosa CB-4. J Integr Agric 13(8):1719–1726
Zermane N, Souissi T, Kroschel JR, Sikora R (2007) Biocontrol of broomrape (Orobanchecrenata Forsk. and Orobanchefoetida Poir.) by Pseudomonas fluorescens isolate Bf7-9from the faba bean rhizosphere. Biocontrol Sci Tech 17(5):483–497
Zhang LH, Zhang JL, Liu YC, Cao ZY, Han JM, Yang J, Dong JG (2013) Isolation and structural speculation of herbicide-active compounds from the metabolites of Pythium aphanidermatum. J Integr Agric 12:1026–1032
Acknowledgements
The authors are grateful to the Council of Scientific and Industrial Research (CSIR), New Delhi, India, and The World Academy of Science (TWAS), Italy, for providing the necessary facilities and opportunity to carry out this research. Special thanks to Mr. Rajul Tomar and the whole staff of Microbial Type Culture Collection and Gene Bank (MTCC), Institute of Microbial Technology, Sector 39A, Chandigarh, India for their contribution to the molecular aspect of this work. Also, I like to appreciate Dr. Adejumo Isaac and Miss Onikanni Olayinka for their input in the statistical analysis.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Adetunji, C.O., Oloke, J.K., Prasad, G. et al. Isolation, identification, characterization, and screening of rhizospheric bacteria for herbicidal activity. Org. Agr. 8, 195–205 (2018). https://doi.org/10.1007/s13165-017-0184-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13165-017-0184-8