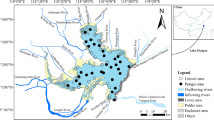
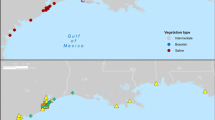
Abstract
Coastal salt marshes are some of the most productive ecosystems on Earth, providing numerous services such as soil carbon storage, flood protection and nutrient filtering, several of which are mediated by the sediment microbiome associated with marsh vegetation. Here, nutrient filtering (nitrate removal through denitrification) was examined by determining microbial (bacteria and archaea) community structure (16S rRNA gene iTag sequencing), diversity, denitrification rates and metabolic potential (assembled metagenomic sequences) in collocated patches of Spartina alterniflora (Spartina) and Juncus roemerianus (Juncus) sediments. The iTag data showed that diversity and richness in Spartina and Juncus sediment microbial communities were highly similar. However, microbial community evenness differed significantly, with the most even communities observed in Juncus sediments. Further, denitrification rates were significantly higher in Juncus compared to Spartina, suggesting oscillations in microbial abundances and in particular the core microbiome identified herein, along with plant diversity influence marsh nitrogen (N) removal. Amplicon and assembled metagenome sequences pointed to a potentially important, yet unappreciated Planctomycetes role in N removal in the salt marsh. Thus, ecosystem perturbations that alter marsh vegetation distribution could impact microbial diversity and may ultimately influence the ecologically important ecosystem functions the marsh sediment microbiome provides.






Similar content being viewed by others
Data Availability
Data is available in the appropriate repositories.
Code Availability
Not applicable.
References
Anderson I, Tobias C, Neikirk B, Wetzel RL (1997) Development of a process-based nitrogen mass balance model for a Virginia (USA) Spartina alterniflora salt marsh: implications for net DIN flux. Marine Ecology Progress Series 159:13–27
Angell JH, Peng X, Ji Q, Craick I, Jayakumar A, Kearns P, Ward BB, Bowen JL (2018) Community composition of nitrous oxide-related genes in salt marsh sediments exposed to nitrogen enrichment. Frontiers in Microbiology 9:1–13
Angermeyer A, Crosby SC, Huber JA (2018) Salt marsh sediment bacterial communities maintain original population structure after transplantation across a latitudinal gradient. PeerJ 6:1–21
Apprill A, McNally S, Parsons R, Weber L (2015) Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microbial Ecology 75:129–137
Awasthi A, Singh M, Soni SK, Singh R, Kalra A (2014) Biodiversity acts as insurance of productivity of bacterial communities under abiotic perturbations. The ISME Journal 8:2445–2452
Battaglia LL, Woodrey MS, Peterson MS, Dillon KS, Visser JM (2012) Wetland Ecosystems of the Northern Gulf Coast. In: Batzer DP, Baldwin, AH (eds). Wetland Habitats of North America: Ecology and Conservation Concerns, pp 75–88. University of California Press.
Bowen JL, Morrison HG, Hobbie JE, Sogin ML (2012) Salt marsh sediment diversity: a test of the variability of the rare biosphere among environmental replicates. ISME Journal 6:2014–2023
Bowen JL, Kearns PJ, Byrnes JEK, Wigginton S, Allen WJ, Greenwood M, Tran K, Yu J, Cronin JT, Meyerson LA (2017) Lineage overwhelms environmental conditions in determining rhizosphere bacterial community structure in a cosmopolitan invasive plant. Nature Communications 8:433
Burke DJ, Hamerlynck EP, Hahn D (2002) Interactions among plant species and microorganisms in salt marsh sediments. Applied and Environmental Microbiology 68:1157–1164
Callahan BJ, Mcmurdie PJ, Rosen MJ, Han AW, Johnson AJA, Holmes SP (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 13:581–583
Callahan BJ, Mcmurdie PJ, Holmes SP (2017) Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME Journal 11:2639–2643
Cao YP, Green PG, Holden PA (2008) Microbial community composition and denitrifying enzyme activities in salt marsh sediments. Applied and Environmental Microbiology 74:7585–7595
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman F, Costello EK, Fierer N, Gonzalez A, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nature Methods 7:335–336
Caporaso JG, Lauber CL, Walters W, Berg-Lyons D, Lozupone C, Turnbaugh PJ, Fierer N, Knight R (2011) Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceedings of the National Academy of Sciences of the United States of America 108(Suppl):4516–22
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, Gormley N, Gilbert JA, Smith G, Knight R (2012) Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME Journal 6:1621–1624
Chao A (1984) Nonparametric estimation of the number of classes in a population. Scandinavian Journal of Statistics 11:265–270
Chapin FS, Walker BH, Hobbs RJ, Hooper DU, Lawton JH, Sala OE, Tilman D (1997) Biotic Control over the Functioning of Ecosystems. Science (80-.) 277:500 LP–504
Chaumeil P-A, Mussig AJ, Hugenholtz P, Parks DH (2019) GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database. Bioinformatics 36(6):1925–1927
Chen I-MA, Chu K, Palaniappan K, Pillay M, Ratner A, Huang J, Huntemann M, Varghese N, White JR, Seshadri R, Smirnova T, Kirton E, Jungbluth SP, Woyke T, Eloe-Fadrosh EA, Ivanova NN, Kyrpides NC (2019) IMG/M v.5.0: an integrated data management and comparative analysis system for microbial genomes and microbiomes. Nucleic Acids Research 47:D666–D677
Cleary DFR, Polónia ARM, Sousa AI, Lillebø AI, Queiroga H, Gomes NCM (2016) Temporal dynamics of sediment bacterial communities in monospecific stands of Juncus maritimus and Spartina maritima. Plant Biology 18:824–834
Cleary DFR, Coelho FJRC, Oliveira V, Gomes NCM, Polónia ARM (2017) Sediment depth and habitat as predictors of the diversity and composition of sediment bacterial communities in an inter-tidal estuarine environment. Marine Ecology 38:1–15
Costanza R, d’Arge R, de Groot R, Farber S, Grasso M, Hannon B, Limburg K, Naeem S, O’Neill RV, Paruelo J, Raskin RG, Sutton P, van den Belt M (1997) The value of the world’s ecosystem services and natural capital. Nature 387:253–260
Costanza R, Groot R, De, Sutton P, Ploeg S, Van Der, Anderson SJ, Kubiszewski I, Farber S, Turner RK (2014) Changes in the global value of ecosystem services. Global Environmental Change 26:152–158
Damashek J, Francis CA (2018) Microbial nitrogen cycling in estuaries: from genes to ecosystem processes. Estuaries and Coasts 41:626–660
Davis DA, Gamble MD, Bagwell CE, Bergholz PW, Lovell CR (2011) Responses of salt marsh plant rhizosphere diazotroph assemblages to changes in marsh elevation, edaphic conditions and plant host species. Microbial Ecology 61:386–398
Dollhopf SL, Hyun J-H, Smith AC, Adams HJ, O’Brien S, Kostka JE (2005) Quantification of ammonia-oxidizing bacteria and factors controlling nitrification in salt marsh sediments. Applied and Environmental Microbiology 71:240–246
Ehrlich P, Wilson E (1991) Biodiversity studies: science and policy. Science (80-.) 253:758 LP – 762
Eisenhauer N, Scheu S, Jousset A (2012) Bacterial diversity stabilizes community productivity. PLoS One 7:1–5
Eleuterius LN (1976) The distribution of Juncus roemerianus in the Salt Marshes of North America. Chesapeake Science 17:289–292
Elsey-Quirk T, Seliskar DM, Gallagher JL (2011) Nitrogen pools of macrophyte species in a Coastal Lagoon Salt Marsh: Implications for seasonal storage and dispersal. Estuaries and Coasts 34:470–482
Elsey-Quirk T, Seliskar DM, Sommerfield CK, Gallagher JL (2011) Salt marsh carbon pool distribution in a mid-Atlantic lagoon, USA: sea level rise implications. Wetlands 31:87–99
Gillies LE, Thrash JC, de Rada S, Rabalais NN, Mason OU (2015) Archaeal enrichment in the hypoxic zone in the northern Gulf of Mexico. Environmental Microbiology 17:3847–3856
Glöckner FO, Kube M, Bauer M, Teeling H, Lombardot T, Ludwig W, Gade D, Beck A, Borzym K, Heitmann K, Rabus R, Schlesner H, Amann R, Reinhardt R (2003) Complete genome sequence of the marine planctomycete Pirellulasp. strain 1. Proceedings of the National Academy of Sciences 100:8298 LP – 8303.
Gribsholt B, Kostka JE, Kristensen E (2003) Impact of fiddler crabs and plant roots on sediment biogeochemistry in a Georgia saltmarsh. Marine Ecology Progress Series 259:237–251
Hamady M, Knight R (2009) Microbial community profiling for human microbiome projects: tools, techniques, and challenges. Genome Research 19:1141–1152
Hamersley MR, Howes BL (2005) Coupled nitrification-denitrification measured in situ in a Spartina alterniflora marsh with a 15NH4 + tracer. Marine Ecology Progress Series 299:123–135
Hopkinson CS, Giblin AE (2008) Nitrogen dynamics of coastal salt marshes. In: Capone DG, Bronk DA, Mulholland MR, Carpenter EJ (eds) Nitrogen in the marine environment. Academic, San Diego, pp 991–1036
Jordan SJ, Stoffer J, Nestlerode JA (2011) Wetlands as sinks for reactive nitrogen at continental and global scales: a meta-analysis. Ecosystems 14:144–155
Joye SB, Hollibaugh T (1995) Influence of sulfide inhibition of nitrification on nitrogen regeneration in sediments. Science 270:623–625
Kang DD, Froula J, Egan R, Wang Z (2015) MetaBAT, an efficient tool for accurately reconstructing single genomes from complex microbial communities. PeerJ 3:e1165
Kang DD, Li F, Kirton E, Thomas A, Egan R, An H, Wang Z (2019) MetaBAT 2: an adaptive binning algorithm for robust and efficient genome reconstruction from metagenome assemblies. PeerJ 7:e7359
Kartal B, Kuypers MMM, Lavik G, Schalk J, op den Camp HJM, Jetten MSM, Strous M (2007) Anammox bacteria disguised as denitrifiers: nitrate reduction to dinitrogen gas via nitrite and ammonium. Environmental Microbiology 9:635–642
Kartal B, Keltjens J, Jetten M (2011) Metabolism and genomics of anammox bacteria. In: Arp Ward B (ed) Nitrification. ASM Press, Washington DC, pp 181–200
King GM, Klug MJ, Wiegert RG, Chalmers AG (1982) Relation of soil water movement and sulfide concentration to Spartina alterniflora production in a Georgia salt marsh. Science (80-) 218:61–63
Koop-Jakobsen K, Giblin AE (2010) The effect of increased nitrate loading on nitrate reduction via denitrification and DNRA in salt marsh sediments. Limnology and Oceanography 55:789–802
Koop-Jakobsen K, Wenzhöfer F (2015) The dynamics of plant-mediated sediment oxygenation in Spartina anglica rhizospheres—a planar optode study. Estuaries and Coasts 38:1–13
Koretsky CM, Haveman M, Cuellar A, Beuving L, Shattuck T, Wagner M (2008) Influence of Spartina and Juncus on saltmarsh sediments. I. Pore water geochemistry. Chemical Geology 255:87–99
Kostka JE, Gribsholt B, Petrie E, Dalton D, Skelton H, Kristensen E (2002) The rates and pathways of carbon oxidation in bioturbated saltmarsh sediments. Limnology and Oceanography 47:230–240
Kuypers MMM, Sliekers AO, Lavik G, Schmid M, Jørgensen BB, Kuenen JG, Sinninghe Damsté JS, Strous M, Jetten MSM (2003) Anaerobic ammonium oxidation by anammox bacteria in the Black Sea. Nature 422:608–611
Ledford TC, Mortazavi B, Tatariw C, Mason OU (2020) Elevated nutrient inputs to marshes differentially impact carbon and nitrogen cycling in two northern Gulf of Mexico saltmarsh plants. Biogeochemistry 149:1–16
Linthurst RA, Seneca ED (1980) The effects of standing water and drainage potential on the Spartina Alterniflora-substrate complex in a North Carolina salt marsh. Estuarine and Coastal Marine Science 11:41–52
Mavrodi OV, Jung CM, Eberly JO, Hendry SV, Namjilsuren S, Biber PD, Indest KJ, Mavrodi DV (2018) Rhizosphere microbial communities of spartina alterniflora and juncus roemerianus from restored and natural tidal marshes on deer Island, Mississippi. Frontiers in Microbiology 9:1–13
Mendelssohn IA, Morris JT (2000) Eco-physiological controls on the productivity of Spartina Alterniflora Loisel. In: Kreeger DA (ed) Concepts and Controversies in Tidal Marsh Ecology. Kluwer Academic, Dordrecht
Meng J, Xu J, Qin D, He Y, Xiao X, Wang F (2014) Genetic and functional properties of uncultivated MCG archaea assessed by metagenome and gene expression analyses. ISME Jounal 8:650–659
Miley GA, Kiene RP (2004) Sulfate reduction and porewater chemistry in a gulf coastJuncus roemerianus(Needlerush) marsh. Estuaries and Coasts 27:472–481
Mitsch WJ, Gosselink JG (2015) Wetlands, Fifth Edition. John Wiley & Sons.
Moffett KB, Gorelick SM (2016) Relating salt marsh pore water geochemistry patterns to vegetation zones and hydrologic influences. WATER Resources Research 52:1729–1745
Neubauer SC (2013) Ecosystem responses of a tidal freshwater marsh experiencing saltwater intrusion and altered hydrology. Estuaries and Coasts 36:491–507
Newell SY (2001) Multiyear patterns of fungal biomass dynamics and productivity within naturally decaying smooth cordgrass shoots. Limnology and Oceanography 46:573–583
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2019) vegan: Community Ecology Package
Oliveira V, Santos AL, Coelho F, Gomes NCM, Silva H, Almeida A, Cunha  (2010) Effects of monospecific banks of salt marsh vegetation on sediment bacterial communities. Microbial Ecology 60:167–179
Oliveira V, Santos AL, Aguiar C, Santos L, Salvador ÂC, Gomes NCM, Silva H, Rocha SM, Almeida A, Cunha  (2012) Prokaryotes in salt marsh sediments of Ria de Aveiro: Effects of halophyte vegetation on abundance and diversity. Estuarine, Coastal and Shelf Science 110:61–68
Parada AE, Needham DM, Fuhrman JA (2015) Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environmental Microbiology 18:1403–1414
Parks DH, Imelfort M, Skennerton CT, Hugenholtz P, Tyson GW (2015) CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Research 25:1043–1055
Parks DH, Chuvochina M, Waite DW, Rinke C, Skarshewski A, Chaumeil P-A, Hugenholtz P (2018) A standardized bacterial taxonomy based on genome phylogeny substantially revises the tree of life. Nature Biotechnology 36:996–1004
Paulson JN, Stine OC, Bravo HC, Pop M (2013) Differential abundance analysis for microbial marker-gene surveys. Nature Methods 10:1200–1202
Pielou EC (1966) The measurement of diversity in different types of biological collections. Journal of Theoretical Biology 13:131–144
Pomeroy LR, Wiegert RG (1981) Ecology of a salt marsh. Springer-Verlag, New York
Revelle W (2018) psych: Procedures for personality and psychological research. https://CRAN.R-project.org/package=psych
Rietl AJ, Overlander ME, Nyman AJ, Jackson CR (2016) Microbial community composition and extracellular enzyme activities associated with Juncus roemerianus and Spartina alterniflora vegetated sediments in Louisiana Saltmarshes. Microbial Ecology 71:290–303
Scheuner C, Tindall BJ, Lu M, Nolan M, Lapidus A, Cheng J, Goodwin L, Pitluck S, Huntemann M, Liolios K, Pagani I, Mavromatis K, Ivanova N, Pati A, Chen A, Palaniappan K, Jeffries CD, Hauser L, Land M et al (2014) Complete genome sequence of Planctomyces brasiliensis type strain (DSM 5305T), phylogenomic analysis and reclassification of Planctomycetes including the descriptions of Gimesia gen. nov., Planctopirus gen. nov. and Rubinisphaera gen. nov. and emended descriptions of the order Planctomycetales and the family Planctomycetaceae. Standards in Genomic Sciences 9:10
Schlesner H (1989) Planctomyces brasiliensis sp. nov., a Halotolerant Bacterium from a Salt Pit. Systematic and Applied Microbiology 12:159–161
Seitzinger, SP, Nielsen, LP, Caffrey, J, Christensen, PB (1993) Denitrification measurements in aquatic sediments: A comparison of three methods. Biogeochemistry 23(3):147–167
Seitzinger S, Harrison JA, Böhlke JK, Bouwman AF, Lowrance R, Peterson B, Tobias C, Van Drecht G (2006) Denitrification across landscapes and waterscapes: a synthesis. Biogeosciences Discussions 16:2064–2090
Shade A, Handelsman J (2012) Beyond the Venn diagram: The hunt for a core microbiome. Environmental Microbiology 14:4–12
Shannon CE, Weaver W (1949) The mathematical theory of communication. The University of Illinois Press, Urbana
Sorensen J (1978) Denitrification rates measured in a marine sediment as measured by acetlyene inhibition technique. Applied and Environmental Microbiology 36:139–143
Sorensen J, Tiedje JM, Firestone RB (1980) Inhibition by sulfide of nitric and nitrous oxide reduction by denitrifying Pseudomonas fluorescens. Applied and Environmental Microbiology 39:105–108
Starr G, Staudhammer CL, Starr G, Jarnigan JR, Staudhammer CL, Cherry JA (2018) Variation in ecosystem carbon dynamics of saltwater marshes in the northern Gulf of. Wetlands Ecology and Management 26:581–596
Stout J (1984) The ecology of irregularly flooded salt marshes of the Northeastern Gulf of Mexico. A Community Profile U.S. Department of the Interior, Fish and Wildlife Service, Washington DC
Strous M, Fuerst JA, Kramer EHM, Logemann S, Muyzer G, van de Pas-Schoonen KT, Webb R, Kuenen JG, Jetten MSM (1999) Missing lithotroph identified as new planctomycete. Nature 400:446–449
Sunagawa S, Coelho LP, Chaffron S, Kultima JR, Labadie K, Salazar G, Djahanschiri B, Zeller G, Mende DR, Alberti A, Cornejo-castillo FM, Costea PI, Cruaud C, Ovidio F, Engelen S, Ferrera I, Gasol JM, Guidi L, Hildebrand F et al (2015) Structure and function of the global ocean microbiome. Science 348(6237):1261359
Thamdrup B, Dalsgaard T (2002) Production of N2 through anaerobic ammonium oxidation coupled to nitrate reduction in marine sediments. Applied and Environmental Microbiology 68:1312–1318
Tiner RW (1984) Wetlands of the United States–Current status and recent trends. U.S. Fish and Wildlife Service Report, Washington, D.C.
Tobias CR, Macko SA, Anderson IC, Canuel EA, Harvey JW (2001) Tracking the fate of a high concentration groundwater nitrate plume through a fringing marsh: A combined groundwater tracer and in situ isotope enrichment study. Limnology and Oceanography 46:1977–1989
Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI (2007) The human microbiome project. Nature 449:804–810
Valiela I, Cole ML (2002) Comparative evidence that salt marshes and mangroves may protect seagrass meadows from land-derived nitrogen loads. Ecosystems 5:92–102
Valiela I, Teal JM (1979) The nitrogen budget of a salt marsh ecosystem. Nature 280:652
Valiela I, Geist M, McClelland J, Tomasky G (2000) Nitrogen loading from watersheds to estuaries: Verification of the Waquoit Bay Nitrogen Loading Model. Biogeochemistry 49:277–293
Velinsky DJ, Paudel B, Quirk T, Piehler M, Smyth A (2017) Salt marsh denitrification provides a significant nitrogen sink in Barnegat Bay, New Jersey. Journal of Coastal Research 78:70–78
Vitousek, P.M., Mooney, H.A., Lubchenco, J., and Melillo, J.M. (1997) Human domination of earth’s ecosystems. Science (80-.) 277:494 LP – 499
Wang J, Vine CE, Balasiny BK, Rizk J, Bradley CL, Tinajero-Trejo M, Poole RK, Bergaust LL, Bakken LR, Cole JA (2016) The roles of the hybrid cluster protein, Hcp and its reductase, Hcr, in high affinity nitric oxide reduction that protects anaerobic cultures of Escherichia coli against nitrosative stress. Molecular Microbiology 100:877–892
Wang M, Yang P, Salles JF, Saleem M (2016) Distribution of root-associated bacterial communities along a salt-marsh primary succession. Frontiers in Plant Science 6:1188
Wiegart RG, Freeman BJ (1990) Tidal marshes of the southeastern Atlantic coast: a community profile. US Fish Wildl Serv 85:1–80
Wiegert RG, Chalmers AG, Randerson PF (1983) Productivity gradients in salt marshes: the response of Spartina alterniflora to experimentally manipulated soil water movement. Oikos 41:1–6
Wilson BJ, Mortazavi B, Kiene RP (2015) Spatial and temporal variability in carbon dioxide and methane exchange at three coastal marshes along a salinity gradient in a northern Gulf of Mexico estuary. Biogeochemistry 123:329–347
Wittebolle L, Marzorati M, Clement L, Balloi A, Daffonchio D, Heylen K, De Vos P, Verstraete W, Boon N (2009) Initial community evenness favours functionality under selective stress. Nature 458:623–626
Wu Y-W, Tang Y-H, Tringe SG, Simmons BA, Singer SW (2014) MaxBin: an automated binning method to recover individual genomes from metagenomes using an expectation-maximization algorithm. Microbiome 2:26
Yachi S, Loreau M (1999) Biodiversity and ecosystem productivity in a fluctuating environment: The insurance hypothesis. Proceedings of the National Academy of Sciences 96:1463–1468
Yilmaz P, Parfrey LW, Yarza P, Gerken J, Pruesse E, Quast C, Schweer T, Peplies J, Ludwig W, Glockner FO (2014) The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Research 42:D643–D648
Zedler JB, Kercher S (2005) WETLAND RESOURCES: Status, trends, ecosystem services, and restorability. Annual Review of Environment and Resources 30:39–74
Acknowledgements
We would like to thank A. Kleinhuizen and L. Linn with assistance in the laboratory. We would also like to thank Philip Hugenholtz and Gene Tyson for the opportunity to analyze metagenomic data at the Australian Centre for Ecogenomics. This project was funded by the National Science Foundation’s Division of Chemical, Bioengineering, Environmental and Transport Systems grants 1438092 and 1643486. Metagenomic sequencing was provided by the Joint Genome Institute through a small-scale community sequencing project grant 503678.
Funding
This project was funded by the National Science Foundation’s Division of Chemical, Bioengineering, Environmental and Transport Systems grants 1438092 and 1643486. Metagenomic sequencing was provided by the Joint Genome Institute through a small-scale community sequencing project grant 503678.
Author information
Authors and Affiliations
Contributions
BM and PC collected samples and determined denitrification rates. LK carried out DNA extractions, library preparation and 16S rRNA gene sequencing. OUM carried out bioinformatics and statistical analyses of the 16S rRNA gene sequence data. OUM and JZ carried out metagenome assembly and analyzed MAGs. OUM and BM wrote the manuscript.
Corresponding author
Ethics declarations
Conflicts of Interest/Competing Interests
The authors declare no conflict of interest.
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
The authors provide consent for publication.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Mason, O.U., Chanton, P., Knobbe, L.N. et al. New Insights Into the Influence of Plant and Microbial Diversity on Denitrification Rates in a Salt Marsh. Wetlands 41, 33 (2021). https://doi.org/10.1007/s13157-021-01423-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13157-021-01423-8