Abstract
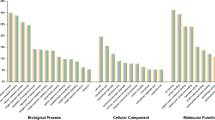
Understanding the effects of inbreeding is critical for the long-term viability of shellfish breeding and for understanding inbreeding depression (ID) of the offspring of selfed individuals and full-sib crosses. The bay scallop (Argopecten irradians) is a simultaneous hermaphroditic marine bivalve with distinct male and female gonads and is a species with external fertilization. It is a canonical model invertebrate for analysis of the genetics of ID and evolution of outcrossing mating system. ID in bivalves is well documented with regard to the phenotype and molecular markers aspects, but little is known on how inbreeding affects genome-wide gene expression. Here, we compare the gene expression profiles of adductor muscle of A. irradians lines inbred to F=0.5 with those of its counterpart outbred lines which are originated from the same paternal individual. Inbred and outbred individuals have some differences in growth traits. The shell length and the total weight of inbred scallops are lower than the counterpart of outbred lines. We found that 1 175 genes were up-regulated while 1 390 were downregulated in inbred lines. Regarding components, differentially expressed genes (DEGs) were concentrated on sarcomeres, contractile filaments, and cell membranes. Among others, whereas regarding their function, the Gene Ontology (GO) was concentrated on transferase, phosphotransferase, peptidase, and others. The DEGs-enriched metabolic pathways mainly belonged to the oxidative phosphorylation pathway and apoptosis pathway. We used another two families to verify our bioinformatic results. The selfed and the outbred family originated from different ancestors, but the selfed family showed obvious ID. By using real-time quantitative reverse transcriptase polymerase chain reaction (qRT-PCR), we verified that DEGs with inbreeding are involved in the oxidative phosphorylation and cell apoptosis pathways, suggesting that inbreeding may affect energy metabolism and homeostasis.
Similar content being viewed by others
References
Bijlsma R, Bundgaard J, van Putten W F. 1999. Environmental dependence of inbreeding depression and purging in Drosophila melanogaster. Journal of Evolutionary Biology, 12(6): 1125–1137, doi: https://doi.org/10.1046/j.1420-9101.1999.00113.x
Birchler J A, Riddle N C, Auger D L, et al. 2005. Dosage balance in gene regulation: biological implications. Trends in Genetics, 21(4): 219–226, doi: https://doi.org/10.1016/j.tig.2005.02.010
Charlesworth B, Hughes K A. 1999. The maintenance of genetic variation in life-history traits. In: Singh R S, Krimbas C B, eds. Evolutionary Genetics: from Molecules to Morphology. Cambridge: Cambridge University Press
Darwin C R. 1876. The Effects of Cross and Self-Fertilization in the Vegetable Kingdom. London: John Murray
DePristo M A, Weinreich D M, Hartl D L. 2005. Missense meanderings in sequence space: a biophysical view of protein evolution. Nature Reviews Genetics, 6(9): 678–687, doi: https://doi.org/10.1038/nrg1672
Fowler K, Whitlock M C. 1999a. The distribution of phenotypic variance with inbreeding. Evolution, 53(4): 1143–1156, doi: https://doi.org/10.1111/evo.1999.53.issue-4
Fowler K, Whitlock M C. 1999b. The variance in inbreeding depression and the recovery of fitness in bottlenecked populations. Proceedings of the Royal Society B: Biological Sciences, 266(1433): 2061–2066, doi: https://doi.org/10.1098/rspb.1999.0887
Hedgecock D, Sly F. 1990. Genetic drift and effective population sizes of hatchery-propagated stocks of the Pacific oyster, Crassostrea gigas. Aquaculture, 88(1): 21–38, doi: https://doi.org/10.1016/0044-8486(90)90316-F
Huang X, Madan A. 1999. CAP3: A DNA sequence assembly program. Genome Research, 9(9): 868–877, doi: https://doi.org/10.1101/gr.9.9.868
Kristensen T N, Dahlgaard J, Loeschcke V. 2002. Inbreeding affects Hsp70 expression in two species of Drosophila even at benign temperatures. Evolutionary Ecology Research, 4: 1209–1216
Kristensen T N, Dahlgaard J, Loeschcke V. 2003. Effects of inbreeding and environmental stress on fitness–using Drosophila buzzatii as a model organism. Conservation Genetics, 4(4): 453–465, doi: https://doi.org/10.1023/A:1024763013798
Kristensen T N, Sørensen P, Kruhøffer M, et al. 2005. Genome-wide analysis on inbreeding effects on gene expression in Drosophila melanogaster. Genetics, 171(1): 157–167, doi: https://doi.org/10.1534/genetics.104.039610
Longwell A C, Stiles S S. 1973. Gamente cross incompatibility and inbreeding in the commercial American oyster, Crassostrea virginica Gmelin. Cytologia, 38(3): 521–533, doi: https://doi.org/10.1508/cytologia.38.521
Mallet A L, Haley L E. 1983. Growth rate and survival in pure population matings and crosses of the oyster Crassostrea virginica. Canadian Journal of Fisheries and Aquatic Sciences, 40(7): 948–954, doi: https://doi.org/10.1139/f83-121
Newkirk G F, Haley L E. 1983. Selection for growth rate in the European oyster, Ostrea edulis: response of second generation groups. Aquaculture, 33(1–4): 149–155
Pletcher S D, Macdonald S J, Marguerie R, et al. 2002. Genome-wide transcript profiles in aging and calorically restricted Drosophila melanogaster. Current Biology, 12(9): 712–723, doi: https://doi.org/10.1016/S0960-9822(02)00808-4
Reed D H, Briscoe D A, Frankham R. 2002. Inbreeding and extinction: the effect of environmental stress and lineage. Conservation Genetics, 3(3): 301–307, doi: https://doi.org/10.1023/A:1019948130263
Saavedra C. 1997. Low effective sizes in hatchery populations of the European oyster (Ostrea edulis): implications for the management of genetic resources. Journal of Shellfish Research, 16: 441–446
Sangster T A, Lindquist S, Queitsch C. 2004. Under cover: causes, effects and implications of hsp90-mediated genetic capacitance. BioEssays, 26: 348–362, doi: https://doi.org/10.1002/(ISSN)1521-1878
Vermeulen C J, Bijlsma R. 2004a. Changes in mortality patterns and temperature dependence of lifespan in Drosophila melanogaster caused by inbreeding. Heredity, 92(4): 275–281, doi: https://doi.org/10.1038/sj.hdy.6800412
Vermeulen C J, Bijlsma R. 2004b. Characterization of conditionally expressed mutants affecting age-specific survival in inbred lines of Drosophila melanogaster: lethal conditions and temperature-sensitive periods. Genetics, 167(3): 1241–1248, doi: https://doi.org/10.1534/genetics.103.023721
Author information
Authors and Affiliations
Corresponding authors
Additional information
Foundation item: The National Natural Science Foundation of China under contract Nos 30800842 and 31502164; the Program of the Chinese Academy of Science under contract No. ZKCX2-211; the Earmarked Fund for Modern Agro-industry Technology Research System under contract No. CARS-49.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Wang, Y., Zhang, S., Zhang, G. et al. Molecular mechanism of inbreeding effects based on RNA-Seq analysis of the adductor muscle of bay scallop (Argopecten irradians). Acta Oceanol. Sin. 38, 44–51 (2019). https://doi.org/10.1007/s13131-019-1351-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13131-019-1351-4