Abstract
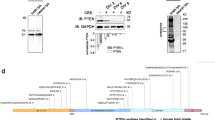
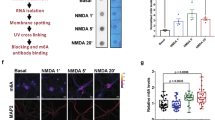
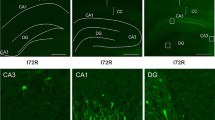
Prolonged translation arrest in post-ischemic hippocampal CA1 pyramidal neurons precludes translation of induced stress genes and directly correlates with cell death. We evaluated the regulation of mRNAs containing adenine- and uridine-rich elements (ARE) by assessing HuR protein and hsp70 mRNA nuclear translocation, HuR polysome binding, and translation state analysis of CA1 and CA3 at 8 h of reperfusion after 10 min of global cerebral ischemia. There was no difference between CA1 and CA3 at 8 h of reperfusion in nuclear or cytoplasmic HuR protein or hsp70 mRNA, or HuR polysome association, suggesting that neither mechanism contributed to post-ischemic outcome. Translation state analysis revealed that 28 and 58 % of unique mRNAs significantly different between 8hR and NIC, in CA3 and CA1, respectively, were not polysome-bound. There was significantly greater diversity of polysome-bound mRNAs in reperfused CA3 compared to CA1, and in both regions, ARE-containing mRNAs accounted for 4–5 % of the total. These data indicate that posttranscriptional ARE-containing mRNA regulation occurs in reperfused neurons and contributes to post-ischemic outcome. Understanding the differential responses of vulnerable and resistant neurons to ischemia will contribute to the development of effective neuroprotective therapies.




Similar content being viewed by others
References
Hossmann K. Disturbances of cerebral protein synthesis and ischemic cell death. Prog Brain Res. 1993;96:161–77.
Nowak TJ. Protein synthesis and the heat shock/stress response after ischemia. Cerebrovasc Brain Metab Rev. 1990;2:345–66.
Nowak TJ. Localization of the 70 kDa stress protein mRNA induction in gerbil brain after ischemia. J Cereb Blood Flow Metab. 1991;11:432–9.
Planas AM, Soriano MA, Estrada A, Sanz O, Martin F, Ferrer I. The heat shock stress response after brain lesions: induction of 72 kDa heat shock protein (cell types involved, axonal transport, transcriptional regulation) and protein synthesis inhibition. Prog Neurobiol. 1997;51:607–36.
Uemura Y, Kowall NW, Beal MF. Global ischemia induces NMDA receptor-mediated c-fos expression in neurons resistant to injury in gerbil hippocampus. Brain Res. 1991;542:343–7.
Roberts GG, Di Loreto M, Marshall M, Wang J, DeGracia DJ. Hippocampal cellular stress responses after global brain ischemia and reperfusion. Antioxidants Redox Signal. 2007;9:2265–75.
DeGracia DJ, Jamison JT, Szymanski JJ, Lewis MK. Translation arrest and ribonomics in post-ischemic brain: layers and layers of players. J Neurochem. 2008;106:2288–301.
Sharp FR, Kinouchi H, Koistinaho J, Chan PH, Sagar SM. HSP70 heat shock gene regulation during ischemia. Stroke. 1993;24:I72–75.
Hu BR, Martone ME, Jones YZ, Liu CL. Protein aggregation after transient cerebral ischemia. J Neurosci. 2000;20:3191–9.
DeGracia DJ, Hu BR. Irreversible translation arrest in the reperfused brain. J Cereb Blood Flow Metab. 2007;27:875–93.
Jamison JT, Kayali F, Rudolph J, Marshall M, Kimball SR, DeGracia DJ. Persistent redistribution of poly-adenylated mRNAs correlates with translation arrest and cell death following global brain ischemia and reperfusion. Neuroscience. 2008;154:504–20.
Jamison JT, Szymanski JJ, Degracia DJ. Organelles do not colocalize with mRNA granules in post-ischemic neurons. Neuroscience. 2011;199:394–400.
Tenenbaum SA, Carson CC, Lager PJ, Keene JD. Identifying mRNA subsets in messenger ribonucleoprotein complexes by using cDNA arrays. Proc Natl Acad Sci. 2000;97:14085–90.
Tenenbaum SA, Lager PJ, Carson CC, Keene JD. Ribonomics: identifying mRNA subsets in mRNP complexes using antibodies to RNA-binding proteins and genomic arrays. Methods. 2002;26:191–8.
Good PJ. A conserved family of elav-like genes in vertebrates. Proc Natl Acad Sci U S A. 1995;92:4557–61.
Ma W-J, Cheng S, Campbell C, Wright A, Furneaux H. Cloning and characterization of HuR, a ubiquitously expressed Elav-like protein. J Biol Chem. 1996;271:8144–51.
Hinman M, Lou H. Diverse molecular functions of Hu proteins. Cell Mol Life Sci. 2008;65:3168–81.
Bakheet T, Williams BRG, Khabar KSA. ARED 3.0: the large and diverse AU-rich transcriptome. Nucl Acids Res. 2006;34:D111–114.
Fan XC, Steitz JA. Overexpression of HuR, a nuclear-cytoplasmic shuttling protein, increases the in vivo stability of ARE-containing mRNAs. EMBO J. 1998;17:3448–60.
Peng S, Chen C-YA XN, Shyu A-B. RNA stabilization by the AU rich element binding protein, HuR, an ELAV protein. EMBO J. 1998;17:3461–70.
Gallouzi IE, Brennan CM, Steitz JA. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. RNA. 2001;7:1348–61.
Mazan-Mamczarz K, Galban S, Lopez de Silanes I, Martindale JL, Atasoy U, Keene JD, et al. RNA-binding protein HuR enhances p53 translation in response to ultraviolet light irradiation. Proc Natl Acad Sci. 2003;100:835–9.
Galbán S, Martindale JL, Mazan-Mamczarz K, de Silanes IL, Fan J, Wang W, et al. Influence of the RNA-binding protein HuR in pVHL-regulated p53 expression in renal carcinoma cells. Mol Cell Biol. 2003;23:7083–95.
Gallouzi I, Brennan CM, Stenberg MG, Swanson MS, Eversole A, Maizels N, et al. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci. 2000;97:3073–8.
Kedersha N, Anderson P. Stress granules: sites of mRNA triage that regulate mRNA stability and translatability. Biochem Soc Trans. 2002;30:963–9.
Keene JD. RNA regulons: coordination of post-transcriptional events. Nat Rev Genet. 2007;8:533–43.
Smith M, Bendek G, Dahlgren N, Rosen I, Wieloch T, Siesjo BK. Models for studying long-term recovery following forebrain ischemia in the rat. 2. A 2-vessel occlusion model. Acta Neuropathol. 1984;69:385–401.
DeGracia DJ, Rudolph J, Roberts GG, Rafols JA, Wang J. Convergence of stress granules and protein aggregates in hippocampal cornu ammonis 1 at later reperfusion following global brain ischemia. Neuroscience. 2007;146:562–72.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-[Delta][Delta]CT method. Methods. 2001;25:402–8.
MacManus JP, Graber T, Luebbert C, Preston E, Rasquinha I, Smith B, et al. Translation-state analysis of gene expression in mouse brain after focal ischemia. J Cereb Blood Flow Metab. 2004;24:657–67.
DeGracia DJ, O’Neil BJ, Frisch C, Krause GS, Skjaerlund JM, White BC, et al. Studies of the protein synthesis system in the brain cortex during global ischemia and reperfusion. Resuscitation. 1993;25:161–70.
Zong Q, Schummer M, Hood L, Morris DR. Messenger RNA translation state: the second dimension of high-throughput expression screening. Proc Natl Acad Sci U S A. 1999;96:10632–6.
Beilharz TH, Preiss T. Translational profiling: the genome-wide measure of the nascent proteome. Brief Funct Genomic Proteomic. 2004;3:103–11.
Simon R, Lam A, Li M-C, Ngan M, Menenzes S, Zhao Y. Analysis of gene expression data using BRB-array tools. Cancer Inform. 2007;3:11–7.
Cartharius K, Frech K, Grote K, Klocke B, Haltmeier M, Klingenhoff A, et al. MatInspector and beyond: promoter analysis based on transcription factor binding sites. Bioinformatics. 2005;21:2933–42.
Brennan CM, Gallouzi I, STEITZ JA. Protein ligands to HuR modulate its interaction with target mRNAs in vivo. J Cell Biol. 2000;151:1–14.
Simon RP, Cho H, Gwinn R, Lowenstein DH. The temporal profile of 72-kDa heat-shock protein expression following global ischemia. J Neurosci. 1991;11:881–9.
Truettner JS, Hu K, Liu CL, Dietrich WD, Hu B. Subcellular stress response and induction of molecular chaperones and folding proteins after transient global ischemia in rats. Brain Res. 2009;1249:9–18.
Van Elzen R, Moens L, Dewilde S. Expression profiling of the cerebral ischemic and hypoxic response. Expert Rev Proteomics. 2008;5:263–82.
Gibbs RA, Weinstock GM, Metzker ML, Muzny DM, Sodergren EJ, Scherer S, et al. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521.
Longo F, Wang S, Narasimhan P, Zhang J, Chen J, Massa SM, et al. cDNA cloning and expression of stress-inducible rat hsp70 in normal and injured rat brain. J Neurosci Res. 1993;36:325–35.
Atasoy U, Watson J, Patel D, Keene J. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111:3145–56.
Antic D, Keene J. Messenger ribonucleoprotein complexes containing human ELAV proteins: interactions with cytoskeleton and translational apparatus. J Cell Sci. 1998;111:183–97.
Fukao A, Sasano Y, Imataka H, Inoue K, Sakamoto H, Sonenberg N, et al. The ELAV protein HuD stimulates Cap-dependent translation in a poly(A)- and eIF4A-dependent manner. Molecular Cell. 2009;36:1007–17.
Büttner F, Cordes C, Gerlach F, Heimann A, Alessandri B, Luxemburger U, et al. Genomic response of the rat brain to global ischemia and reperfusion. Brain Res. 2009;1252:1–14.
Mehra A, Lee KH, Hatzimanikatis V. Insights into the relation between mRNA and protein expression patterns: I. Theoretical considerations. Biotechnol Bioeng. 2003;84:822–33.
Yanagihara T. Cerebral anoxia: effect on neuron-glia fractions and polysomal protein synthesis. J Neurochem. 1976;27:539–43.
Kogure K, Kato H. Altered gene expression in cerebral ischemia. Stroke. 1993;24:2121–7.
Yagita Y, Sakoda S. Kitagawa K [Gene expression in brain ischemia]. Brain Nerve. 2008;60:1347–55.
Stenzel-Poore MP, Stevens SL, King JS, Simon RP. Preconditioning reprograms the response to ischemic injury and primes the emergence of unique endogenous neuroprotective phenotypes: a speculative synthesis. Stroke. 2007;38:680–5.
VanGilder RL, Huber JD, Rosen CL, Barr TL. The transcriptome of cerebral ischemia. Brain Res Bull. 2012;88:313–9.
Hai T, Wolford CC, Chang Y-S. ATF3, a hub of the cellular adaptive-response network, in the pathogenesis of diseases: is modulation of inflammation a unifying component? Gene Expr. 2010;15:1–11.
Cherry J, Karschner V, Jones H, Pekala PH. HuR, an RNA-binding protein, involved in the control of cellular differentiation. In Vivo. 2006;20:17–23.
Voronina E, Seydoux G, Sassone-Corsi P, Nagamori I. RNA granules in germ cells. Cold Spring Harb Perspect Biol. 2011;3:a002774.
Fan J, Yang X, Wang W, Wood WH, Becker KG, Gorospe M. Global analysis of stress-regulated mRNA turnover by using cDNA arrays. Proc Natl Acad Sci U S A. 2002;99:10611–6.
Abdelmohsen K, Lal A, Kim HH, Gorospe M. Posttranscriptional orchestration of an anti-apoptotic program by HuR. Cell Cycle. 2007;6:1288–92.
Acknowledgments
This work was sponsored by NIH NINDS Grant No. NS057167 (D.J.D.), an NINDS Ruth Kirsten F30 Predoctoral Fellowship Grant No. NINDS NS063651 (J.J.S.), and a Thomas C. Rumble Fellowship, Wayne State University (J.T.J.).
Compliance with Ethics Requirements
All institutional and national guidelines for the care and use of laboratory animals were followed.
Conflict of Interest
All authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
(PDF 723 KB)
Online Resource 2
(PDF 463 KB)
Rights and permissions
About this article
Cite this article
Szymanski, J.J., Wang, H., Jamison, J.T. et al. HuR Function and Translational State Analysis Following Global Brain Ischemia and Reperfusion. Transl. Stroke Res. 4, 589–603 (2013). https://doi.org/10.1007/s12975-013-0273-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12975-013-0273-2